Protein Research
Proteins have diverse functions in biological phenomena such as catalysis for biological reactions, response to stimuli, and molecular transport. Fujifilm Wako offers standard reagents such as protein extraction reagents and SDS-PAGE reagents, as well as unique reagents such as "Phos-tag™" products for separation and detection of phosphoproteins.
Product Line-up
More Information
What Are Proteins?
Proteins are polymers of amino acids linked by peptide bonds. Proteins make up about 15% of the components of cells and play a central role in biological phenomena. According to the central dogma of molecular biology, genetic information in DNA is transcribed into RNA, which is translated into the final product, a protein. Proteins have a variety of structures depending on the sequence of amino acids, and the structures determine their functions. Their roles are highly diverse and include catalyzing chemical reactions in living organisms, transmitting and receiving signals, and providing mechanical support and movement to cells and tissues.
Structure and Function of Proteins
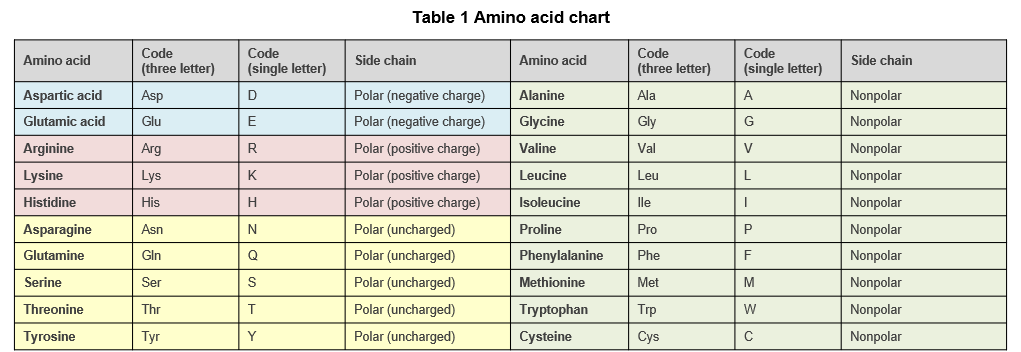
Proteins in living organisms are composed of 20 different amino acids (Table 1), and the amino acid sequence is the main factor that determines the structure of the protein. The α-helix and β-sheet are common secondary structures found in proteins. In addition, the distribution of polar and non-polar amino acids, the presence or absence of disulfide bonds formed between cysteine side chains, and post-translational modifications such as phosphorylation influence the three-dimensional structure of proteins.
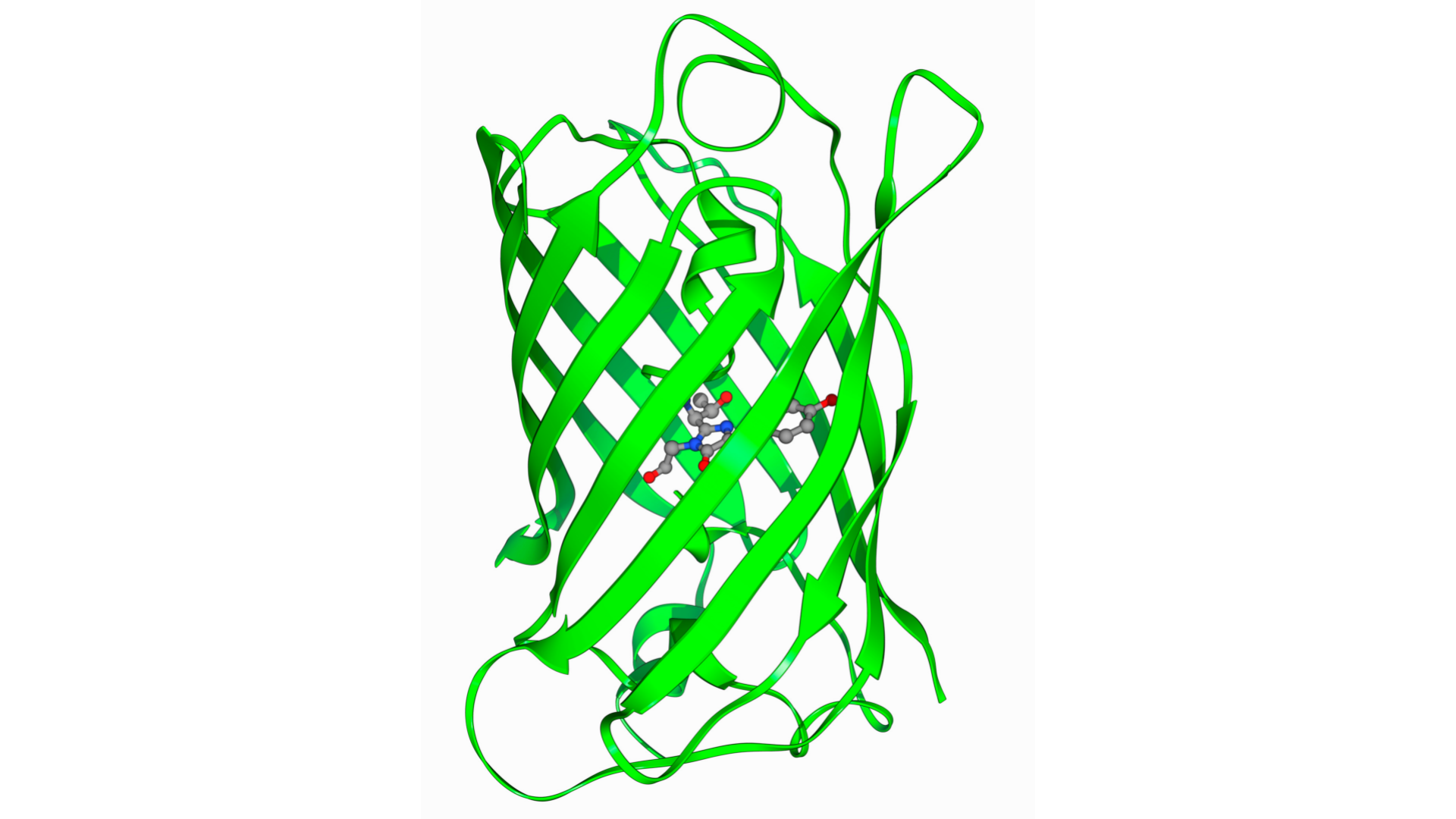
Due to their three-dimensional structures, proteins can bind specifically to certain molecules, called ligands. Protein-ligand binding occurs primarily through relatively weak bonds such as hydrogen bonds, van der Waals forces, and hydrophobic interactions.
Enzymes are a type of protein that specifically bind to ligands called substrates, and catalyze specific chemical reactions. The three-dimensional structure of the enzyme determines its substrates and the chemical reactions. When an enzyme undergoes modification, such as phosphorylation, or when another molecule is bound to the enzyme, the three-dimensional structure changes, and the activity of the enzyme changes accordingly.
Structure and Function of Proteins
To understand a protein, it is necessary to know what kind of protein it is (identification), what kind of structure it has (structural analysis), and what kind of function it has (functional analysis). The methods commonly used in protein research are described below.
Expression
If the gene encoding a target protein is already known, the protein can be produced in cells by introducing a protein expression vector containing a cDNA of the target protein. An appropriate vector needs to be selected depending on the type of cells to be transfected, whether transient or stable expression is desired, and whether tags should be added.
Extraction and Purification
To extract proteins from cells and tissues, the cells must be lysed or crushed. Generally, cell membranes are either lysed in a lysis buffer containing a detergent, or physically crushed. To extract membrane proteins, detergents that solubilize membrane proteins (membrane protein solubilizers) are used.
Column chromatography is often used for protein purification. Based on the properties of the protein to be purified, appropriate methods are selected such as ion exchange chromatography, gel filtration chromatography, and affinity chromatography. If the protein of interest has no ligand that can be used for affinity chromatography, an affinity tag (epitope tag) can be added to the protein and purified using the ligand for the affinity tag.
Separation and Identification
Protein electrophoresis is an effective method to separate specific proteins from a protein mixture. SDS-PAGE is the most widely used electrophoretic separation method. In SDS-PAGE, proteins are given negative charges by addition of sodium dodecyl sulfate (SDS) and then electrophoresed on a polyacrylamide gel. SDS-PAGE can separate proteins by molecular weight.
Proteins separated by SDS-PAGE or other methods can be identified by mass spectrometry. A piece of gel containing the target protein is excised, and the protein is digested in the gel by proteases (peptidases). The peptide fragments are then recovered from the gel and subjected to mass spectrometry (LC-MS/MS or MALDI-TOF MS) to determine the mass of each fragment. The mass data can be searched against a database to identify the protein.
Detection and Quantification
To determine the presence or absence of a target protein or its expression level, a molecule that binds specifically to the protein is used. Antibodies are frequently used for detection and quantification of proteins due to their high specificity. Methods of detection of proteins with antibodies includes western blotting and immunostaining. In western blotting, proteins are separated by SDS-PAGE and transferred to a membrane for antigen-antibody reaction. In immunostaining, antibodies are added to tissues or cells for antigen-antibody reaction. ELISA can accurately quantify a target protein in samples. In ELISA, target proteins are detected using plates coated with immobilized antigens or antibodies and enzyme-linked antibodies.
When evaluating the expression level of a target protein, the total protein content in different samples should be the same to allow for proper comparison. The BCA protein assay, Bradford assay, Lowry assay, Pyrogallol Red Molybdate assay, etc. are available for quantification of total protein with relatively simple procedures.
Analysis of Post-translational Modifications
Post-translational modifications of proteins have significant impact on protein structure and function. Among them, protein phosphorylation is frequently used to control protein activity in cells. Phospho-specific antibodies are often used to examine the phosphorylation state of target proteins. Phos-tag™ is a functional molecule that can trap phosphoproteins and can be used to separate, purify, analyze, and detect phosphoproteins without the use of phospho-specific antibodies. Phos-tag™, developed by the Functional Molecular Science Laboratory, Graduate School of Biomedical and Health Sciences, Hiroshima University, has become a useful tool for the analysis of phosphoproteins.
References
- “Protein Experiment Handbook” ed. by Takenawa, T., Yodosha, Japan, (2003). (Japanese)
- Alberts, B. et al.: “Essential Cell Biology 2nd ed.”, Garland Science/Taylor & Francis Group, (2003).
- “Protein Experiment Protocol vol.1” ed. by Okada, M. and Miyazaki, K., Yodosha, Japan, (2011). (Japanese)
- “Protein Experiment Protocol vol.2” ed. by Okada, M., Miki, H. and Miyazaki, K., Yodosha, Japan, (2011). (Japanese)
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.




