Genetics
Genetics has been significantly contributing to advances in the field of life science and have been creating diagnostic and treatment techniques, food, and energies, etc., which currently provide the basis for a better life for humankind. Fujifilm Wako provides several reagents for genetics.
Product Line-up
-
- DNA Extractor® Kit Series
- [NIPPON GENE] DNA Extraction Reagents (Animal Tissue / Cell / Blood / Hair)
- [NIPPON GENE] DNA Extraction Reagents (Plant / Food)
- [NIPPON GENE] DNA Extraction Reagent (Soil, Fecal)
- Disposable Homogenizer - BioMasher Series
- [NIPPON GENE] DNA Extraction Reagents (Plasmid DNA)
- [NIPPON GENE] DNA Extraction Reagents (Agarose)
- [NIPPON GENE] DNA Purification Reagents (PCR Product)
- Proteinase K
- Coprecipitants
More Information
What is DNA?

Deoxyribonucleic acid (DNA) is a compound that records and transmits genetic information of an organism, and is composed of 2'-deoxyribose (five-carbon sugar), phosphoric acid, and base. The bases of DNA consist of four types: adenine (A) and guanine (G), which have a purine skeleton, and cytosine (C) and thymine (T), which have a pyrimidine skeleton. Adenine and thymine, and guanine and cytosine, can form complementary base pairs through hydrogen bonding.
The smallest unit of nucleic acid in DNA is a deoxyribonucleotide, in which a base is linked to the 1'-position and a phosphate is linked to the 5'-position of 2'-deoxyribose. Deoxyribonucleotides are linked together in a straight line by a phosphate ester bond between a phosphate at the 5'-position and a hydroxyl group at the 3'-position of deoxyribose to form a long polynucleotide chain. Two polynucleotide chains that are assembled so that they serve as templates for each other face each other in an antiparallel position to form double-stranded DNA in a double-helical structure.
Alignment of bases, called the base sequence, comprises the main body of genetic information. A sequence of three bases is called a codon, and one codon encodes one amino acid. DNA is read from the 5' to 3' direction in the synthesis of encoded amino acids.
What is RNA?
Ribonucleic acid (RNA) is a nucleic acid formed by linking ribonucleotides, composed of ribose (five-carbon sugar), phosphate, and base. The bases of RNA consist of four types: adenine (A) and guanine (G), which have a purine skeleton, and cytosine (C) and uracil (U), which have a pyrimidine skeleton. Adenine and uracil, and guanine and cytosine, can form complementary base pairs through hydrogen bonding.
RNA is mainly involved in the transfer and expression of genetic information, and there are various types of RNA, including messenger RNA, in which genetic information of DNA is transcribed; transfer RNA, which acts as an adaptor connecting a messenger RNA with an amino acid; and ribosomal RNA, which constitutes ribosomes essential for translation.
RNA usually exists in a single-stranded form and does not have the limitation of double helix structure that DNA has. Therefore, depending on the base sequences, RNA can locally form a double-stranded structure such as a stem loop, and in some cases, can form a more complicated three-dimensional structure. RNA with a higher order structure can have catalytic activity, and RNA that acts as an enzyme in cells is called a ribozyme. Representative ribozymes include RNase P, which cleaves tRNA from the precursor RNA.
Difference between DNA and RNA
Although deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are very similar, they differ in the following ways:
(1) The sugar used for the main chain of DNA is 2'-deoxyribose, whereas that of RNA is ribose with a hydroxyl group at the 2'-position.
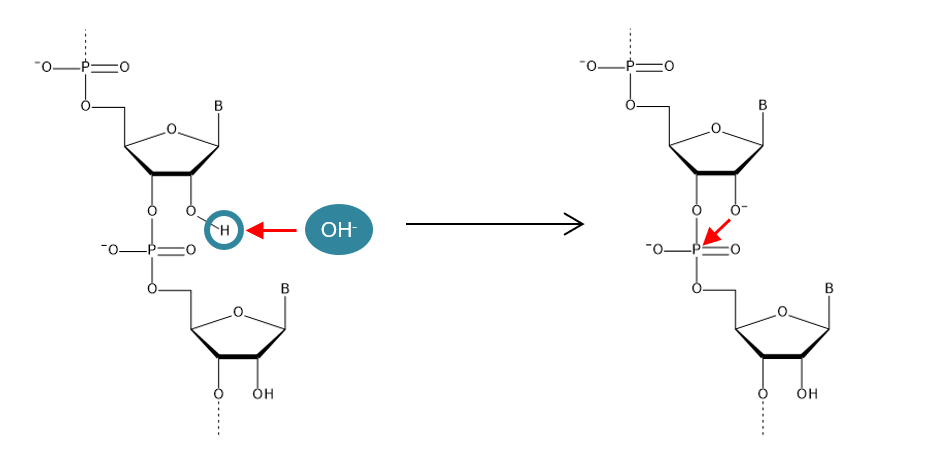
A hydroxyl group at the 2'-position greatly affects the stability of nucleic acids. It is generally considered that RNA is less stable than DNA, and the reason for this is the existence of the 2'-hydroxyl group. A proton in the 2'-hydroxyl group is easily extracted by a basic substance. The deprotonated oxygen atom attacks the phosphorus atom in the phosphodiester bond and breaks the bond between ribonucleotides (Figure 1). An RNA degradation enzyme (RNase) that degrades RNA by a similar mechanism also exists. RNA is easily degraded under mild basic conditions or in the presence of RNase.
A modified nucleotide, in which the 2'-hydroxyl group is changed to an O-methyl group (-OMe), has a five-membered ring structure that is of a C3’-endo type (the 5'-and 3'-carbons protrude in the same direction) similar to that of a ribonucleotide, and is not susceptible to hydrolysis, so it is commonly used in fields such as nucleic acid medicine.
(2) DNA uses thymine as a base, while RNA uses uracil instead of thymine.
DNA and RNA use different bases, thymine and uracil, respectively. However, thymine and uracil have very similar structure, differing only in the presence or absence of the methyl group at the 5'-position, and thus thymine can also be called 5'-methyluracil. Both thymine and uracil can form a complementary bond with adenine.
It is considered that the reason why uracil is not used in DNA is that uracil is a base that is naturally formed by deamination of cytosine. If uracil is used in DNA as a base, the uracil produced by deamination of cytosine cannot be discriminated from naturally occurring uracil. If DNA does not contain uracil, the DNA repair mechanism can recognize uracil produced by deamination of cytosine as an abnormal base and replace it with cytosine.
Types and Functions of RNA
Various types of RNA exist in cells, and each plays an important role. Types and functions of RNA are described below.
▼Messenger RNA (mRNA)
mRNA encodes sequence information for a protein, which is transcribed from a gene (DNA) by RNA polymerase. In eukaryotic cells, after being transcribed from DNA in the nucleus, RNA undergoes processing, including RNA capping, polyadenylation, and splicing. After processing, mature mRNA is transported out of the nucleus and translated into protein. It is said that approximately 1% to 5% of the total RNA is mRNA in a typical mammalian cell.
RNA does not need to transfer to the cell nucleus like DNA, and is less likely to be integrated into the genome. In addition, RNA is easier to produce and modify than proteins are. Therefore, RNA is attracting attention in the fields of nucleic acid medicine and vaccines. Although low stability and immunogenicity when administered to organisms have been issues, the use of modified bases and capping systems (Cap1) and the development of drug delivery systems have led to the practical application of RNA.
▼Transfer RNA (tRNA)
tRNA acts as an adaptor connecting a codon with an amino acid specified by the codon. tRNA has a clover leaf structure, in which an anticodon region complementary to a codon of mRNA, and a region to which amino acids corresponding to the codon are bound are present. tRNA accounts for approximately 15% to 20% of total RNA in mammalian cells.
▼Ribosomal RNA (rRNA)
rRNA is a component of ribosomes, a translation machinery, together with ribosomal proteins. rRNA accounts for approximately 80% to 85% of the total RNA in typical mammalian cells, and the presence of 5S/5.8S/18S/28S rRNA in eukaryotes and 5S/16S/23S rRNA in prokaryotes has been determined from the sedimentation rate under centrifugal force. rRNA can have a three-dimensional structure and act as a ribozyme, catalyzing peptide bond formation between amino acids during protein translation.
▼MicroRNA (miRNA)
miRNA is a type of small-molecule functional RNA, and consists of approximately 22 bases. miRNA has been reported to function as a guide molecule to regulate gene expression at the post-transcriptional level and to have various biological functions. More than 1,000 types of miRNA are thought to be encoded in humans and mice, and research on the elucidation of functions of new miRNA and the search for disease-related miRNA is actively progressing worldwide. In addition, the presence of miRNA not only in cells but also in body fluids such as blood has been reported, and miRNA is attracting attention as a clinical diagnostic marker of diseases such as cancer.
Other than miRNA, there are other functional small-molecule RNAs, including endogenous small interfering RNA (siRNA) and PIWI-interacting RNA (piRNA), which consist of bases of slightly differing numbers. As different AGO proteins bind to them, they can be isolated by immunoprecipitation using these binding proteins.
Other RNA
Functional small-molecule RNAs that has been known since long ago include small nuclear RNA (snRNA), which exists in the nucleus and is involved in RNA splicing, and small nucleolar RNA (snoRNA), which exists in the nucleolus and is involved in rRNA modification.
History of genetic engineering experiments
The double-helical structure was proposed by J. Watson and F. Crick in 19531). It was shown that each of the two helices connected in opposite directions is arranged so that a base is on the inside and a phosphate, bound to deoxyribose, is on the outside, that if a base of one helix is A, its paired base is always T, and if a base of one helix is G, its paired base is always C, and that the opposing bases are linked by hydrogen bonds to stabilize the helical structure. These structural characteristics implied a mechanism for the replication of genetic information and suggested that DNA contained genetic information. The central dogma was proposed by F. Crick in 1958 2), and the flow of genetic information was clarified, in which genetic information is transcribed from DNA to mRNA and is expressed as a protein through translation.
After that discovery, DNA ligase, which ligates DNA fragments, was isolated by B. Weiss and C. Richardson in 1966, and restriction enzymes that recognize and cleave specific nucleotide sequences (palindromic structures) in genes were discovered by W. Arber in Switzerland and H. Smith in the United States in 1968. P. Berg of Stanford University then succeeded in a gene recombination experiment in which DNA of a carcinogenic virus, SV40, was introduced into Escherichia coli in 1972, and S. Cohen and H. Boyer developed gene recombination technology for Escherichia coli using plasmids in the following year.
With a sense of crisis about the progress of gene recombination technology, P. Berg and other key researchers held the Asilomar conference in California in 1975 to discuss the potential risks of gene recombination and to provide basic guidelines. By the early 1980s, genetic engineering had been established and many genes had been cloned. In 1982, human insulin was approved as the world's first recombinant pharmaceutical product.
In 1983, a polymerase chain reaction (PCR) method was devised as a new gene-amplification method and made it easier to amplify target genes. Automation of PCR was realized using Taq DNA polymerase, discovered in 19763), and was popularized immediately as a general experimental technique. As a result, application of PCR to DNA testing and gene diagnosis/treatment was started 4, 5). The Nobel Prize in Chemistry 1993 was awarded to K. Mullis for the invention of PCR.
In 1983, the year in which the PCR method was devised, A. Feinberg and B. Vogelstein reported the presence of DNA hypomethylation in colorectal cancer tissues 6), suggesting that epigenetics is involved in the development of human cancers. In 1987, R. Holliday reported an association between DNA methylation and epigenetics in oncogenesis and aging 7). These results led to research on epigenetics, in which changes in regulation and transmission of gene expression states can occur without change in DNA sequences.
In the 1990s, large-scale genome analysis technology using sequencing techniques was established, and genome projects of many organisms advanced. The human genome project was initiated in 1990 and was completed in 2003. In 2005, a next generation sequencer (NGS) that can perform millions of sequencing reactions simultaneously was put to practical use. NGS enables the analysis of large-scale genome sequences in a short time and plays a major role in the fields of precision medicine, diagnosis and treatment of hereditary diseases, and pharmaceuticals. In addition, NGS brought about the development of metagenome analysis technology, and allowed the progress of microbiota (microbiome) analysis.
Currently, as post-genome research, larger-scale and more complicated analyses, such as transcriptomics, proteomics and bioinformatics are performed. The appearance of genome editing technology, such as ZFN, TALEN, and CRISPR-Cas9, has enabled easier gene modification at the single-base level.
References
- Watson, J. D. and Crick, F. H.: Nature, 171(4361), 964(1953).
Genetical implications of the structure of deoxyribonucleic acid - Crick, F. H. : Symp. Soc. Exp. Biol., 12(138-63), 8(1958).
On protein synthesis - Chien, A., Edgar, D. B. and Trela, J. M.: J. Bacterial., 127(3), 1550(1976).
Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus - Saiki, R. K. et al.: Science, 230(4732), 1350(1985).
Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia - Mullis, K. B. and Faloona, F. A.: Methods Enzymol., 155, 335(1987).
Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction - Feinberg, A. P. and Vogelstein, B.: Nature, 301(5895), 89 (1983).
Hypomethylation distinguishes genes of some human cancers from their normal counterparts - Holliday, R.: Science, 238(4824), 163(1987).
The inheritance of epigenetic defects - “Principles and protocols for nucleic acid experiments by purpose” ed. by Hirao, I., Kurumizaka, H., Yodosha, Japan, (2011). (Japanese)
- “non-coding RNA textbook” ed. by Siomi, M., Nakagawa, S., Asahara, H., Yodosha, Japan, (2015). (Japanese)
- Watson, J. D. et al.: “Molecular Biology of the Gene 7th ed.”, Pearson, United Kingdom, (2013).
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.



