microRNA
microRNA and Ago-Subfamily Proteins
microRNA (miRNA) are small functional RNA molecules (about 22 nucleotides) which act as posttranscriptional regulators of gene expression. They serve many functions within biological organisms1), and more than 1,000 types are known to exist in humans and mice. Worldwide there are many efforts to identify new types of microRNA with unknown functions and elucidate functions of those involved in disease. Recently it has been reported that microRNA exist not only within cells, but also blood and other bodily fluids. They are now garnering attention as possible clinical markers for cancer and other diseases2)。
We offer a full selection of antibodies for various human and mouse Ago-subfamily proteins, as well as immunoprecipitation kits for them. These tools allow comprehensive analysis of microRNA and mRNA contained in RISC in cells and tissue, as well as Ago-bound microRNA in blood.
In cells, microRNA pass through many steps before being incorporated into a multiprotein complex called a RISC (RNA-induced silencing complex). After binding with the main component of Agosubfamily proteins, they bind with mRNA targets and either cleave them or inhibit translation, thus regulating gene expression (Fig. 1)3,4,5). The Ago-subfamily of proteins are characterized by containing both a PAZ domain and a PIWI domain (Fig. 2). There are four types in humans (hAgo1 to hAgo4), and although each expression levels differ from each them in each cell lines (Fig. 3)6). The most commonly expressed Ago-subfamily protein is Ago2, which has unique "Slicer" activity which directly cleaves target RNA. Thus, Ago2 is considered to play an important role in microRNA pathways7, 8, 9,10,11,12,13). Anti-Ago antibodies are used for immunoprecipitation of RISC, allowing recovery of both microRNA and their mRNA targets. This immunoprecipitation method is vital for elucidating the various functions of microRNA14,15,16,17,18,19, 20, 21).
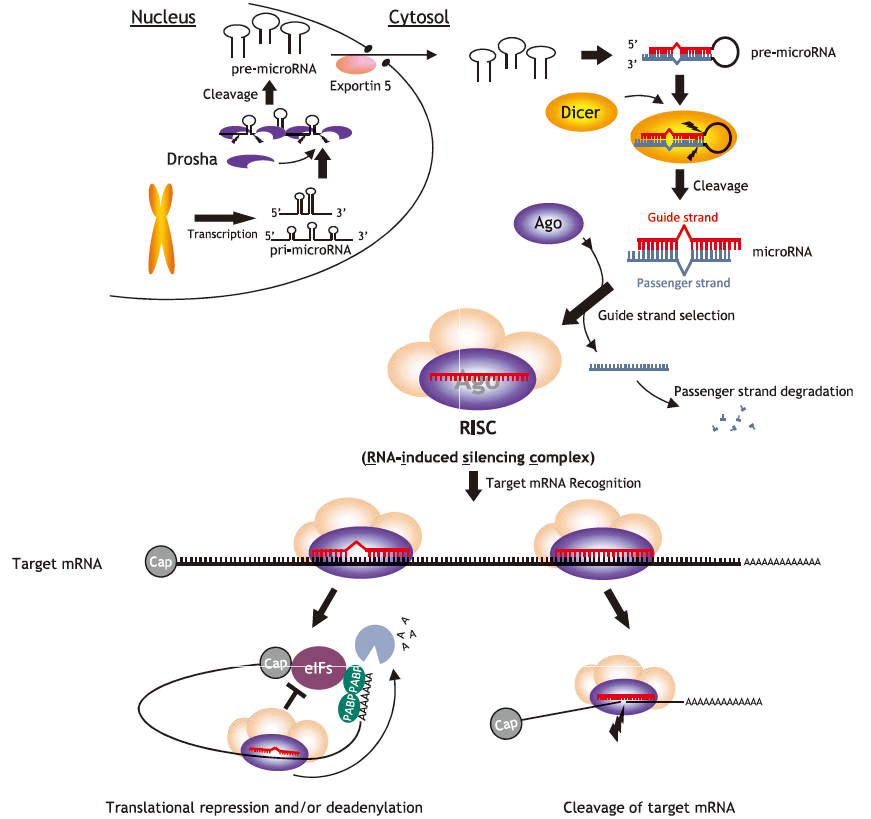
-
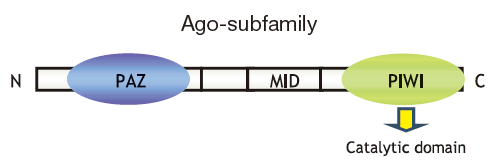
Figure 2 PAZ domain and PIWI domain -
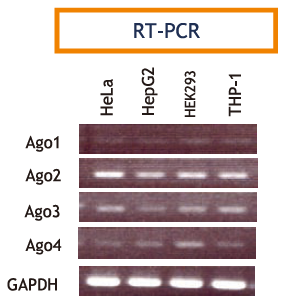
Figure 3 Expression comparison of hAgo1-hAgo4 -

Figure 4 Localization of Ago2
References
- Bartel DP: MicroRNA:genomics, biogenesis, mechanism, and function. Cell, 116:281-297(2004).
- Cortez MA, et al.: MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther, 9:703-711(2009)
- Hammond SM, et al.: An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404:293-296(2000).
- Hammond SM, et al.: Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293:1146-1150(2001).
- Gregory RI, et al.: Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell, 123:631-640(2005).
- Sasaki T, et al.: Identification of eight members of the Argonaute family in the human genome. Genomics, 82:323-330(2003).
- Pillai RS, et al.: Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA, 10:1518-1525(2004).
- Meister G, et al.: Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell, 15:185-197(2004).
- Liu J, et al.: Argonaute2 is the catalytic engine of mammalian RNAi. Science, 305:1437-1441(2004).
- Song JJ, et al.: Crystal structure of Argonaute and its implications for RISC slicer activity. Science, 305:1434-1437(2004).
- Meister G, et al.: Mechanisms of gene silencing by double-stranded RNA. Nature, 431:343-349(2004).
- Hutvagner G, et al.: A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297:2056-2060(2002).
- Yekta S, et al: MicroRNA-directed cleavage of HOXB8 mRNA. Science, 304:594-596(2004).
- Beitzinger M, et al.: Identification of human microRNA targets from isolated Argonaute protein complexes. RNA Biol, 4:76-84(2007).
- Karginov FV, et al.: A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA, 104:19291-19296(2007).
- Easow G, et al.: Isolation of microRNA targets by miRNP immunopurification. RNA, 13:1198-1204(2007).
- Hendrickson DG, et al.: Systematic identification of mRNAs recruited to Argonaute2 by specific microRNAs and corresponding changes in transcript abundance. PloS ONE, 3:e2126(2008).
- Landthaler M, et al.: Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA, 14:2580-2596(2008).
- Chi SW, et al.: Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature, 460:479-486(2009).
- Wang WX, et al.: Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA, Dec.30(2009).
- Hayashida Y, et al.: A useful approach to total analysis of RISC associated RNA. BMC Res Notes, 2:169(2009).
More Information
microRNA Experiment Flow
We offer a microRNA Isolation Kit series containing monoclonal antibodies for various Ago-subfamily proteins and immunoprecipitation methods (Ago IP) for each of these. We also offer tools such as the ISOGEN® series for purifi cation of total RNA from samples or the microRNA Extractor® SP Kit, all allowing purification of fractions containing microRNA from samples depending on research needs. Finally, we offer additional powerful tools to support microRNA research, such as tools to clone cDNA from purifi ed microRNA or target mRNA, electrophoresis tools, and microarray analysis services.
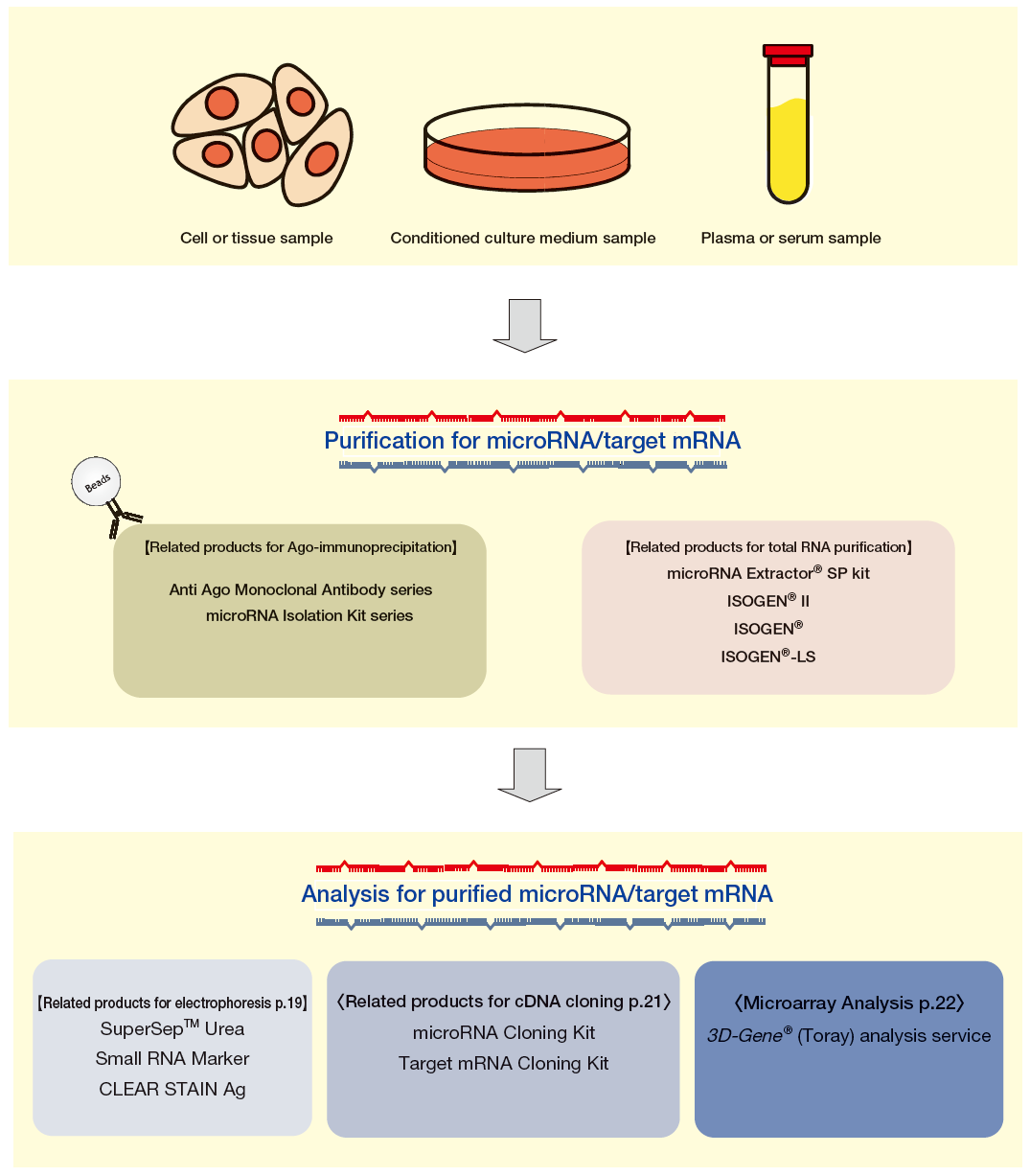
Summary of Ago Immunoprecipitation
We offer a full selection of human and mouse antibodies for Ago-subfamily proteins as well as the microRNA Isolation Kit series based on immunoprecipitation (Ago IP) using these antibodies. These tools offer the following benefi ts over traditional total RNA purifi cation.
Benefi t 1 Concentration of Ago-bound microRNA
Improvement of detection sensitivity of microRNA incorporated in RISC and suppress detection of non-specifi c detection of microRNA not incorporated in RISC.
Example: Comparing populations of Ago IP RNA fractions and total RNA fractions from HeLa cells
Starting with HeLa cells, we used a micro-array (3D-Gene®, Toray) to compare populations of RNA obtained with immunoprecipitation from monoclonal antibodies specifically recognizing each Ago subfamily protein (Ago1 to Ago4) (Ago IP RNA fraction) with microRNA contained in a total RNA fraction prepared using ISOGEN (AGPC) (Fig. 1). Results showed that more microRNA was concentrated in each Ago IP RNA fraction than the total RNA fraction (excluding the Ago4 IP RNA fraction), yielding a stronger signal (Fig. 2). One signal was strongly detected in the total RNA fraction while remaining largely absent in the Ago IP RNA fraction, suggesting that microRNA not incorporated into RISCs were non-specifi cally detected in the total RNA fraction.
-
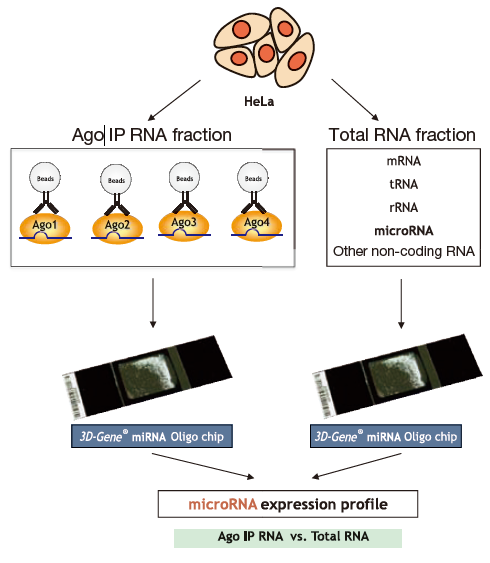
Figure 1. Experiment flow -
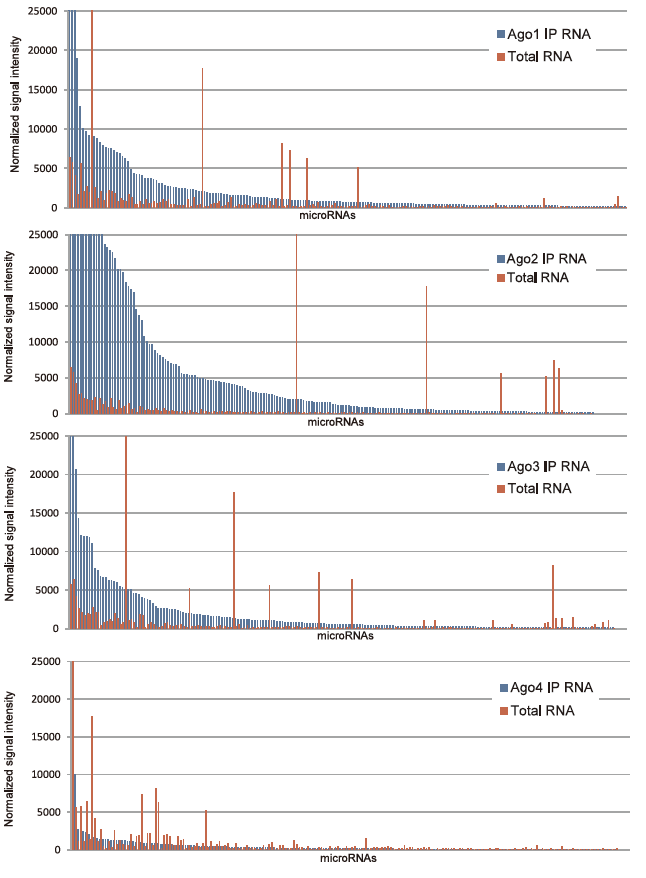
Figure 2. microRNA signal strength comparison between each Ago IP RNA fraction and total RNA fraction
Benefit 2 Concentration of RISC-binding target mRNA
RNA fractions obtained by Ago IP method are applicable to analysis of target mRNA by cDNA cloning, qPCR, microarray, Next-generation DNA sequencer because target mRNA is concentrated.
Example: Concentration of target mRNA of liver-specific microRNA(miR-122)
Either miR-122 or the control of luciferace siRNA (Luc siRNA) were delivered to HepG2, a liver cancer cell line with low expression of miR-122, then Ago2 IP RNA (using anti-Ago2 monoclonal antibody 4G8) and total RNA (using ISOGEN) purified from both samples. Quantitative PCR was then used to compare (Fig. 1) quantities of Aldo A mRNA as a miR-122 target (normalized with GAPDH mRNA) in both cell types. Results show that the quantity of Aldo A mRNA decreased by RISC cleavage, in the total RNA fraction while Aldo A mRNA in the Ago2 IP RNA fraction was concentrated (Fig. 2).
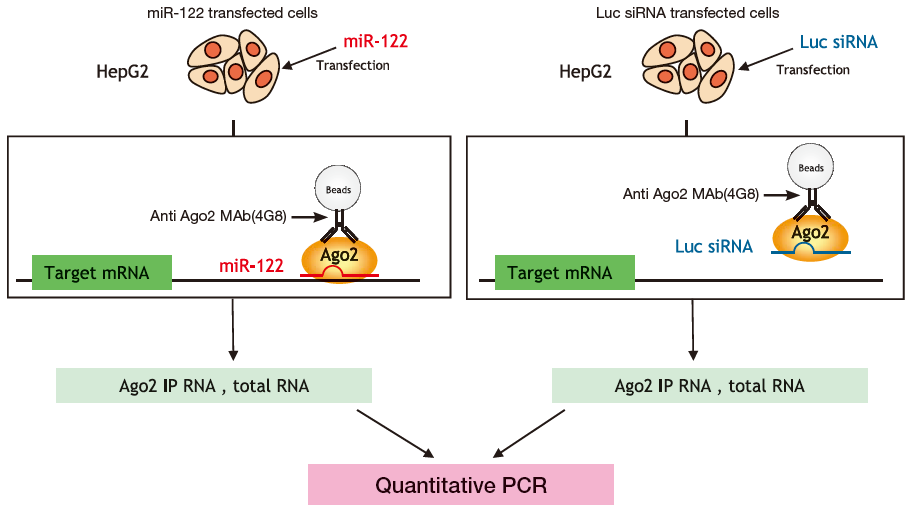
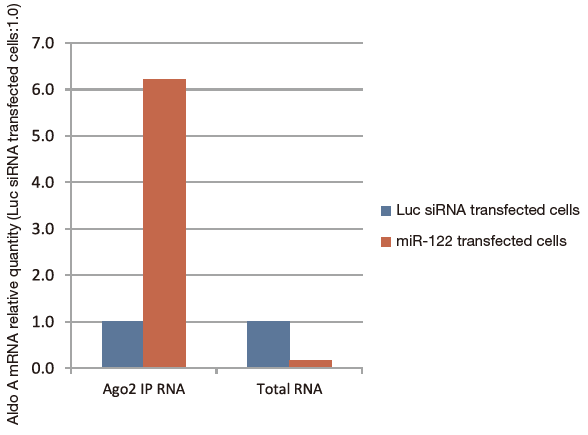
Benefit 3 Concentration of free Ago-bound microRNA from blood samples
Blood is known to include microRNA contained in exosomes, HDL-bound microRNA, and Ago-bound microRNA. Ago immunoprecipitation method allows for specific isolation of Ago-bound microRNA, and it can be used for biomarker searches and other uses.
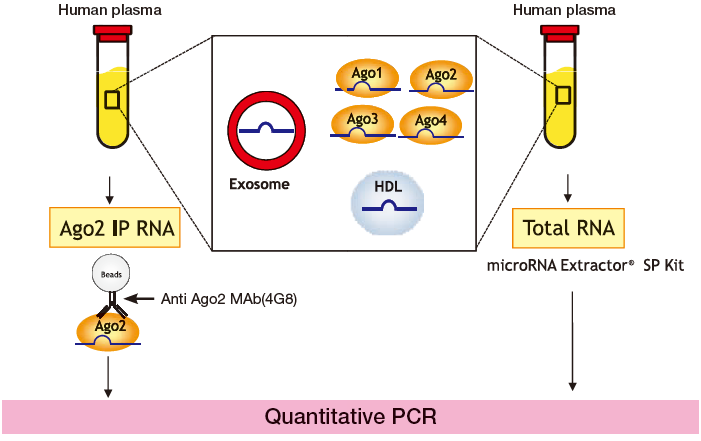
Example 1: Isolation of Ago2-bound microRNA from human plasma
Quantitative PCR (Ct value) was used to measure 12 types of arbitrarily-selected microRNA in pooled plasma of healthy individuals from immunoprecipitated Ago2 IP RNA (using anti-Ago2 monoclonal antibody 4G8) and total RNA (using microRNA Extractor® SP Kit) (Fig. 1). Comparison reveals that most microRNA detected in the total RNA sample was also detected in the Ago2 IP RNA fraction, while types such as miR-22 and miR-92a were highly concentrated in the Ago2 IP RNA fraction (low Ct value compared to total RNA) (Fig. 2).
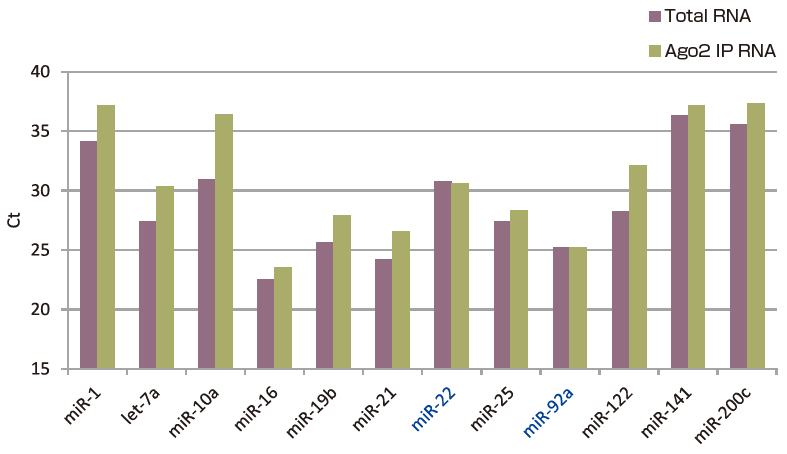
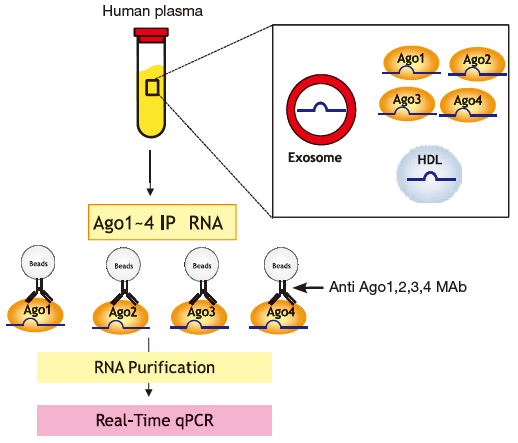
Example 2: Isolation of each Ago-bound microRNA from human plasma
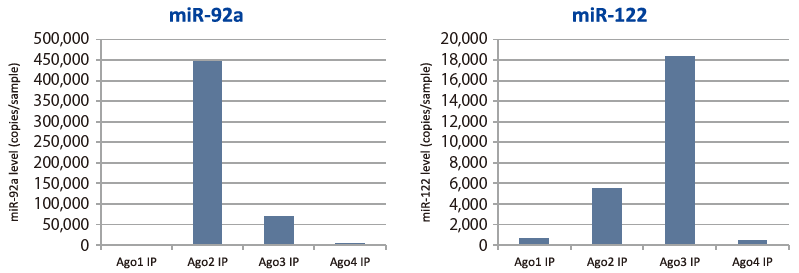
Quantitative PCR was used to compare microRNA (miR-92a and miR-122) contained in immunoprecipitated RNA (Ago 1, 2, 3, 4 IP RNA) using anti-Ago (Ago1 to Ago4) monoclonal antibodies from pooled plasma of healthy individuals (Fig. 1). Results show microRNA present in other Ago IP RNA fractions in addition to the Ago2 IP RNA fraction, with liver-specific miR-122 highly concentrated in the Ago3 IP RNA fraction (Fig. 2).
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.



