Proteome analysis of exosomes
This article was written by Koji Ueda Project for Personalized Cancer Medicine, Cancer Precision Medicine Center, Japanese Foundation for Cancer Research, Japan.
Exosomes are secretory microvesicles derived from intracellular vesicle trafficking pathways, which are involved in the extracellular release of unwanted molecules or delivery of molecules to distant cells. Exosomes are characterized by a lipid bilayer membrane with diameters in the range of several tens to hundreds of nanometers. Specific protein groups (tetraspanin family, Rab family, Tsg101, and Alix) and miRNAs are abundant in exosomes compared to the ratios of cellular composition. However, the biochemical definition of exosomes has not been clearly defined yet. Indeed, a multi-omics analysis based on the particle size of extracellular vesicles revealed that the molecular composition of exosomes differs remarkably depending on the particle size 1). In spite of uncertain definition of exosomes, functional analyses using different exosome purification techniques (exosome samples with different purities) have been conducted worldwide, which for instance reported involvement of exosomes in cancer metastasis, invasion, angiogenesis, and immune cell control.
On the other hand, it is a fact that the molecular profiles of pathogenic cells can be read from pathogenic cell-derived exosomes released into body fluids such as peripheral blood. Moreover, there has been an increase in the commercial use of exosomes for diagnosis. A test (ExoDx® Prostate IntelliScore) to simultaneously detect three RNAs specific to prostate cancer--PCA3 non-coding RNA, ERG mRNA, and SDEF mRNA--from urine exosomes has been developed in a CLIA (Clinical Laboratory Improvement Amendments) laboratory. Now an order as LDT (Laboratory Developed Tests: in vitro diagnostic tests designed, manufactured, and used within one laboratory) to Exosome Diagnostics company is internationally available2). This company has also developed a technology to isolate exosomal DNA/RNA and cfDNA (circulating free DNA) from plasma in one step, and introduced a test to detect EGFR-T790M mutation in non-small cell lung cancer from blood samples (ExoDx® EGFR T790M). This mutation causes drug resistance in approximately 60% patients with lung cancer who are treated with EGFR inhibitors; therefore, prompt diagnosis of acquired resistance is considered to be important in determining transition to effective drugs for EGFR mutations such as osimertinib. A comparative study of 210 patients with lung cancer carrying the same mutation was performed using an approved diagnostic method with cfDNA (cobas® EGFR Mutation Test v2) or the ExoDx® EGFR T790M test. The sensitivity and specificity of T790M detection using cobas® EGFR Mutation Test v2 were 58% and 80%, respectively, while those using the ExoDx® EGFR T790M test were 92% and 89%, respectively3).
These results suggest that tools that allow easy isolation of exosomes with high purity and reproducibility without inter-facility differences are essential for biological researches and clinical studies. In particular, exosomal proteins purified from serum and plasma samples require very high levels of purification efficiency in studies targeting them. Serum and plasma contain more than 60 mg/mL proteins, including high molecular weight complexes, such as lipoprotein. Moreover, it is difficult to remove serum lipoproteins and several free proteins (IgM, α2-macroglobulin, and complement components) even after ultracentrifugal sedimentation, which is a commonly used exosome purification method. Therefore, when serum proteins purified by ultracentrifugal sedimentation are subjected to comprehensive quantitative proteomic analysis by LC/MS, serum-free proteins account for the maximum number and only few exosome-derived proteins are detectable4).
Several exosome purification methods, such as affinity purification using antibodies, polymer precipitation, gel filtration, and direct recovery from resected tissue immersion solution5) are used in addition to ultracentrifugal sedimentation. However, the MagCaptureTM Exosome Isolation Kit PS (hereafter referred to as the MagCapture Kit) is based on affinity purification targeting phosphatidylserine (PS)--a lipid that constitutes the exosome membrane--and offers several advantages over other methods. The binding between Tim-4 protein immobilized on magnet beads and PS requires calcium ion. Based on this characteristic, highly pure exosomes can be isolated by elution using a chelator without eluting non-specific proteins bound to the beads. In addition, this method is effective for exosome isolation from unlimited volume of low-concentration samples directly by magnet beads. Furthermore undenatured elution is a considerably important feature, allowing wide range of subsequent applications, such as particle counting, electron microscopy, and cell administration experiments.
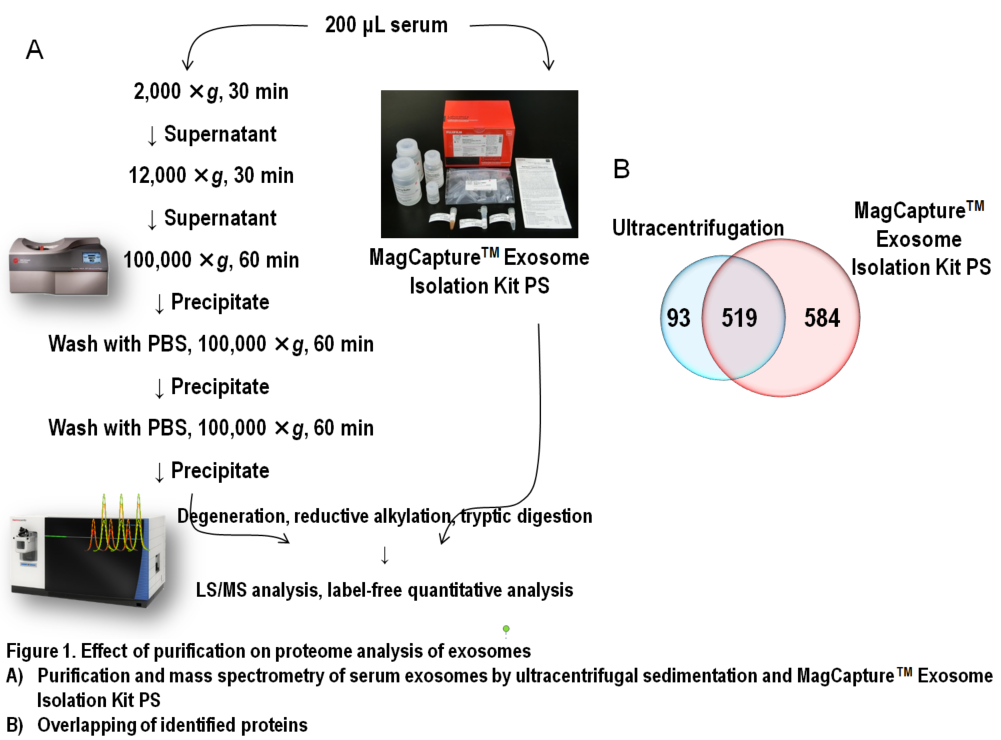
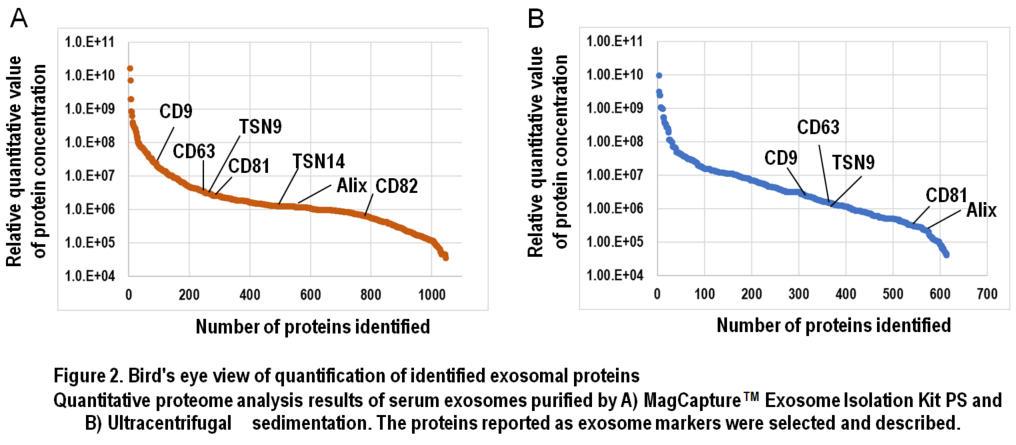
In Figure 1, results of comprehensive proteome analysis for serum exosomes using Orbitrap Fusion Lumos mass spectrometry (Thermo Scientific) are shown. A total of 612 exosomal proteins were identified from exosomes purified with ultracentrifugation, while 1,103 proteins were identified from those purified with the MagCapture kit, providing 1.8-fold greater number of protein identification (Figure 1B). As mentioned previously, ultracentrifugal sedimentation did not remove several free proteins in the serum, which resulted in masking trace exosome-derived signals. Additionally, we found that both the number of exosome marker proteins and their relative concentrations were higher in the exosomes purified by the MagCapture kit than those purified by ultracentrifugation (Figure 2). This suggests that the purity of exosomes obtained using the kit is higher than that using the existing ultracentrifugal sedimentation method.
The use of highly purified exosomes is advantageous and reliable in elucidating the functions of exosomes and exploring the targets of diagnostic and therapeutic drugs. In this regard, the MagCapture kit can serve as the reference point for all exosome studies from the viewpoints of purity, versatility, and reproducibility. However, the products recovered by the kit are a collection of size-independent structures with PS-rich membrane components and should be carefully evaluated and defined by future omics analysis to determine whether they are equivalent to the exosomes referred to in other existing studies. With the development of purification methods including the MagCapture kit as well as methods to detect them, we are expecting to further advance in elucidation of exosomes and their clinical uses.
References
- Zhang, H. et al. :"Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation", Nat. Cell Biol., 20, 332(2018).
- McKiernan, J. et al . :"A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy", JAMA. Oncol., 2, 882(2016).
- Castellanos-Rizaldos, E. et al . :"Exosomebased Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients", Clin. Cancer Res ., DOI : 10.1158/1078-0432. CCR-17-3369(2018).
- Ueda, K.: "The proteomic profile of exosome as a resource of biomarkers", Cell Technology, 32,71(2013).
- Jingushi, K. et al. :"Extracellular vesicles isolated from human renal cell carcinoma tissues disrupt vascular endothelial cell morphology via azurocidin", Int. J. Cancer , 142, 607(2018).




