Quantitative detection of residual DNA with DNA Extractor® Kit
This article was written by Hiroki Fukuchi, Life Science Research Laboratories of FUJIFILM Wako Pure Chemical Corporation, Japan.
Introduction
Since many biopharmaceuticals, including vaccines, are manufactured from cultured cells or microorganisms such as Escherichia coli, it is pointed out that host cell-derived DNA may remain in resultant drug substances and products. Given the possibility that the residual DNA may transfer host cell- or virus-derived oncogenes and that viral DNA may cause infectious events, quantitative detection of residual DNA is important testing for manufacturing and as part of testing such as process validation for biopharmaceuticals. As indicated by a report recommending that up to 100 pg of residual DNA per dose is acceptable for biopharmaceuticals,1) there is increasing need for quantitative detection of residual DNA from host cells as a quality test not only in the US and European countries but also in other countries. To detect traces of residual DNA, it is necessary to extract traces of residual DNA from a sample at a high yield. This article is intended to introduce the usefulness of DNA Extractor® Kit in detecting traces of residual DNA as a DNA purification reagent.
DNA Extractor® Kit (#295-50201, FUJIFILM Wako Pure Chemical Corporation)
Prior to detection and quantification of total DNA in a biologics, it is necessary to separate and purify DNA in the biologics from other biological components such as proteins. In general, DNA is isolated through protease-mediated digestion of specimen followed by extraction with phenol and/or chloroform. However, this method has disadvantages that deleterious phenol and chloroform have to be used and that time- and labor-consuming extraction is required, although quite highly pure DNA can be obtained. In addition, solid-phase extraction via silica carriers, which results in loss of DNA due to adsorption on carriers, is not ideal to recover traces of DNA. Extraction with organic solvents is also not favorable with inefficient recovery of traces of DNA, which is a key issue to be addressed with DNA extraction reagents.
DNA Extractor® Kit (Product code: 295-50201), which we launched in 1992, resolves the aforementioned limitations by using sodium iodide to extract highly pure DNA at high yields through simple procedures.
Principle of DNA Extractor® Kit
The components of the kit* contain sodium iodide and surfactants, which act as protein solubilizers (chaotropic ions) by solubilizing proteins and other biological components in specimens to precipitate (coprecipitate) nucleic acids (mainly DNA) and glycogen selectively when 2-propanol is added.2) Purification steps are simplified to produce precipitates without carriers or organic solvents, unlike the aforementioned methods, enabling the extraction of traces of DNA at high yields.
* Components of the kit:
Sodium Iodide Solution, Sodium N-Lauryl Sarcosinate Solution, Washing Solution (A),
Washing Solution (B), Glycogen Solution
Example of DNA extraction with DNA Extractor® Kit and total DNA quantification
A report on yields of DNA in the presence of diluents or excipients with the use of the kit, which was originally published on page 28 in Wako Pure Chemical Jiho Vol. 60 No. 3 (1992), is introduced here. In this experiment, model solutions were prepared by dissolving usual or excessive doses of substances often used as diluents or excipients (e.g., arginine, urea) or proteins (BSA and human γ-globulin) in phosphate-buffered saline, and a given amount (pg) of calf thymus DNA was added to each model solution. Subsequently, DNA was extracted from 400 μL of model solution according to the instructions on the label of DNA Extractor® Kit. The obtained precipitates were dissolved in 500 μL of phosphate-buffered saline, and total DNA was quantified using Threshold® Total DNA assay system*,3,4) to measure the yield of DNA. Measurement results are presented in Table 1.
Table 1. Yields of DNA in the presence of diluents or excipients
| Diluent and its amount | Amount of DNA (pg) | Yield of DNA (%) | |
|---|---|---|---|
| Sorbitol | 200 mg/mL 50 mg/mL |
50 50 |
95 94 |
| Maltose | 400 mg/mL 200 mg/mL 50 mg/mL |
50 50 50 |
89 102 98 |
| Mannitol | 200 mg/mL | 50 | 84 |
| Dextrose | 200 mg/mL | 50 | 81 |
| Sucrose | 200 mg/mL | 50 | 92 |
| Urea | 1 mol/L | 50 | 106 |
| Arginine | 200 mg/mL | 50 | 110 |
| γ-globulin | 60 mg/mL 60 mg/mL 60 mg/mL |
10 5 2.5 |
102 84 88 |
| BSA | 200 mg/mL | 10 | 95 |
For all five sugars at one or more concentrations, 80% to 110% of DNA was recovered. For arginine and urea, DNA was extracted at a high yield. For both BSA and human γ-globulin, the proteins tested, the yield of DNA was high (some proteins in specimens may precipitate, but the precipitation can be managed with dilution or proteolytic enzymes5)). The aforementioned results show that the kit can extract residual DNA from various samples at high yields in a highly reproducible manner.
* Threshold® Total DNA assay system is manufactured by Molecular Devices for total DNA quantification and designed to quantitatively measure DNA as a molecule rather than as a gene. This system has a detection sensitivity of 2 pg/assay for total DNA.
Example of extraction of traces of residual DNA with DNA Extractor® Kit and DNA quantification by qPCR assay
As mentioned above, Threshold® Total DNA assay system is intended to quantify total DNA, but not to detect any specific gene. Considering the advantages of qPCR quantification for detecting a specific gene, which can be used to detect residual DNA clearly derived from host cells more sensitively than total DNA quantification with Threshold® Total DNA assay system, it was investigated whether traces of DNA could be extracted from potential host cells.
In this experiment, genomic DNA from CHO cells or Escherichia coli, which are widely used to produce proteins and antibodies, was used as a residual DNA model to extract and quantify traces of residual DNA using a qPCR assay. The experimental procedure is presented in Figure 1.
Figure 1. Experimental procedure
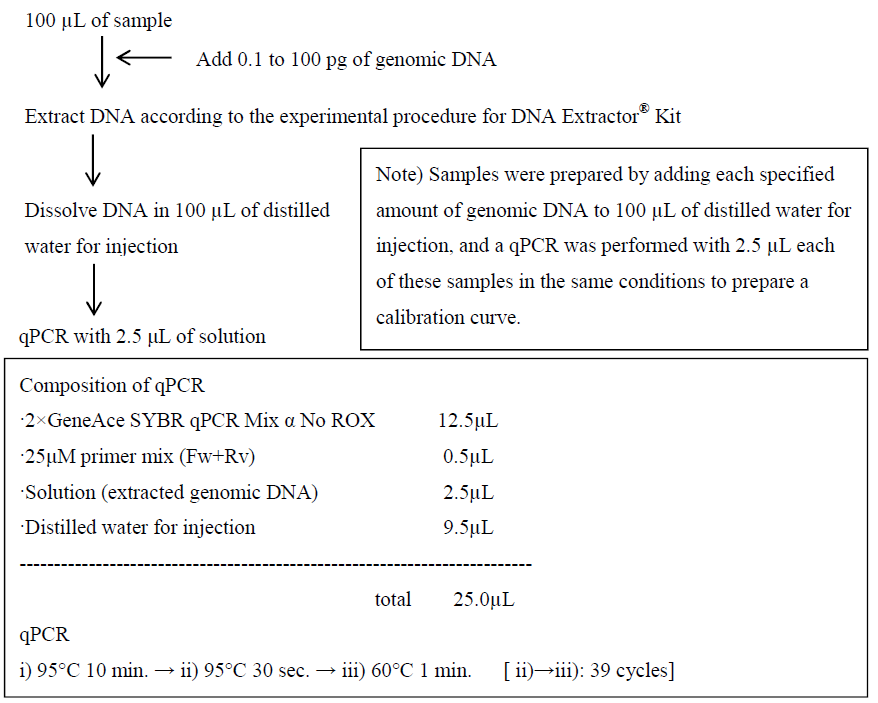
First, DNA was extracted from samples spiked with 0.1 to 100 pg of genomic DNA in 100 μL of water (distilled water for injection) using the kit and then dissolved in 100 μL of water. Subsequently, a qPCR assay was performed to calculate the yield of DNA from a calibration curve prepared simultaneously. The yield of DNA was 85% to 120% for Escherichia coli genome in the range of 1 to 100 pg and for CHO cell genome in the range of 0.1 to 100 pg (the yield was almost 100% up to 1,000 pg for both genomes, although no data are presented here).
Another experiment was performed based on the assumption that residual DNA was contained in a cell culture supernatant. After Panc-1, a human pancreatic cancer-derived cell line, was cultured in 10% FBS DMEM for 3 days, 0.1 pg of genomic DNA from CHO cells was added to 500 μL of culture supernatant to prepare a cell culture supernatant sample. The yield was calculated to be 0.093 pg from a calibration curve. This experiment showed that 0.1 pg (100 fg) of residual DNA in a 500-μL sample, as small as trace in fg units, can be extracted at a high yield using the kit. In addition, traces of residual DNA was recovered from all samples, including water, phosphate-buffered saline, and cell culture supernatant, at as high yields as almost 90% or more (Tables 1 and 2), suggesting that the kit can extract traces of residual DNA from various samples at high yields, as mentioned in the section of total DNA quantification.
Table 2. Yields of genomic DNA
| Sample (added genomic DNA) |
Amount of DNA | Amount of recovered DNA determined from calibration curve | Yield |
|---|---|---|---|
| Distilled water for injection (Escherichia coli) |
0.1pg | ND (less than lower limit of detection) | - |
| 1pg | 1.031pg | 103% | |
| 10pg | 11.09pg | 111% | |
| 100pg | 85.1pg | 85% | |
| Distilled water for injection (CHO cells) |
0.1pg | 0.933pg | 93% |
| 1pg | 1.187pg | 119% | |
| 10pg | 11.57pg | 116% | |
| 100pg | 87.1pg | 87% | |
| Human cell culture supernatant (CHO cells) | 0.1pg | 0.0928pg | 93% |
Conclusion
Residual DNA was recovered from samples at high yields using the kit. DNA extraction from samples is a very important step for quantifying residual DNA, and the kit can extract traces of residual DNA at high yields. In addition, since the kit can be used in various samples and may be useful in extracting residual DNA not only from CHO cells but also from other host cells such as Escherichia coli and yeast, the kit is expected to be used for testing of biopharmaceuticals in future.
References
- Knezevic et al, Biologicals 36:203-211 (2008)
- Ishizawa, M et al, Nucleic Acids Res., 19, 5792 (1991)
- Kung. V. T et al, Anal. Biochem. 187, 220-227 (1990)
- Mizusawa S, Honma R, Pharm Tech. Japan 7, 309-314, 426-431 (1991)
- Wako Pure Chemicals Jiho Vol. 61 No. 1 p. 27 (1993)




