Co-culture studies to decode 'biological PINs' and exosomes
Introduction
Recently, it has become evident that extracellular vesicles, including exosomes, play a major role in the communication of various cells in biology. As such, there has been a dramatic increase in the number of articles on this topic. The current mainstream of exosome research is a method for extracting and analyzing exosomes thought to be related to various diseases and is one of the methods expressed as Liquid Biopsy. However, exosomes are key players in cell-cell interactions, and to truly analyze the functions and disease characteristics of cells via exosomes, apart from a physical fitness measurement-like study where exosomes are extracted and analyzed, it is necessary to perform an interaction study where a phenomenon that occurs in vivo, as in a practice match, is reproduced even in vitro.
Fig. 1 Conceptual diagram of
a biological PIN
This is because, fundamental studies aimed at functional analysis related to exosomes are, broadly speaking, commonly those where, for example, an exosome extracted from cancer cells is administered to other cells, and the behavior of those cells is then observed and analyzed. Then, the functions of that exosome are discussed, followed by the analysis of the inclusions present in that exosome. The functions of the inclusions are further analyzed using molecular biology procedures. The merit of such a method is simply a basic functional analysis, and in case of a machine, one simply pushes the switch and observes what happens. But such a research method is simply pressing a switch, and does the same thing happen in a living organism when a large amount of extracted exosomes is administered? In other words, it is not clear whether such a strong switch is being pressed. Moreover, this study method has a 'security code-like system' (Figure 1) present in the living body, and when the strength, order, etc., are strongly related, it becomes absolutely useless.
The details have been omitted as we plan to publish this article; however, we have confirmed that a 'biological PIN-like system' does exist in the living body. A study where an exosome is extracted and administered to other cells is useful for analyzing the function of a single switch. Henceforth also, there will be no change in the fact that it is a basic analysis method. However, co-culture studies that enable observation of spontaneous interactions between cells without involving extraction techniques are useful for analyzing 'biological PINs.' Although many studies using the co-culture technique have been conducted to date, relatively few studies on exosomes currently use that technique. The use of the co-culture technique is important for analyzing the essential interactions of exosomes. For detailed explanations on exosomes, one can read many reviews on them. This article explains the caution points in exosome study procedures and the co-culture system as an explanatory tool for the purpose of decoding the code numbers in the living body.
Cautions on exosome research
The extraction and analysis of exosomes present in biological samples such as blood and serum culture cells has been the mainstream procedure in exosome research to date, and there are many reports such as those on Liquid Biopsy 1-5). The analysis of functional substances present in exosomes has led to plenty research on these as disease markers, treatment targets, and treatment applications. In particular, miRNA is present in exosomes, and since it was shown to likely migrate between cells6,7), there has been much research on the analysis of miRNA in exosomes, and there have been many reports stating that diseases can be screened using it. Studies on exosomes are commonly conducted using 3 main approaches. (1) Exosomes are extracted, and the inclusions analyzed, (2) the extracted exosomes are administered to other cells, etc., and the reaction confirmed, and (3) the exosomes are labeled and their behavior in cells analyzed. As such, they are studies where (1)-(3) are combined. When classified more simply, they can be broadly categorized into whether or not exosomes are extracted.
The important points here are that in experiments where exosomes are extracted, purified, added to other cells, and their intracellular uptake and changes in those cells are analyzed, 1) attention needs to be paid to the characteristics of the exosome population as per the method used for their extraction, and 2) it is not clear whether the phenomenon caused by the large amount of extracted exosomes simulates the situation in vivo.
1) Effects of extraction methods
When analysis is performed by extracting exosomes, the collected exosomes may form a group with a biased nature depending on the extraction method. This is because various extraction methods are often discussed with regard to their recovery efficiency, but it is important to note that only those exosomes whose properties were affected by the extraction method can be gathered.
Exosome extraction methods include size fractionation using ultracentrifugation, or fractionation by structures with filters and micropores, differences in extraction speed using gel filtration, proteins expressed on the surface, separation by antibodies, and affinity substances using tetraspanin, etc. As such, there is a flood of methods for extracting exosomes.
The reason for the flood of extraction methods is because we do not know what exactly exosomes are. According to early reports, it was thought that the extracellular vesicles containing exosomes secreted from blood platelets and erythrocytes were cellular waste 8,9). In 1983, Dr. Johonstone named vesicles secreted from sheep reticulocytes as exosomes10). Many cells secrete exosomes and are also present in body fluids such as blood. Exosomes have many unknown functions and contain proteins and nucleic acids. They have tetraspanins, a family of membrane proteins, with a structure that penetrates cell membranes with a lipid bilayer four times, membrane transporter proteins, and adhesive molecules such as integrins. In addition to exosomes, extracellular vesicles secreted by cells include plasma membrane-derived microvesicles that are slightly larger in size than exosomes and apoptotic bodies that are released when cells undergo apoptosis11). There was also a period of confusion because the definition of terminology was not uniform among researchers, but in recent years the concept of exosomes has been unified primarily by the International Society of Extracellular Vesicles. Extracellular vesicles are currently divided into three main groups. (1) Exosomes containing a lipid bilayer 50-150 nm in diameter, (2) microvesicles of vesicles secreted directly from cells 100-1000 nm in size, and (3) apoptotic bodies generated from cells that have undergone apoptosis11). In particular, as a breakthrough event, miRNA and RNA were included in exosomes from 2006 to 2007, and the possibility of their movement between cells via exosomes was shown6,7). Since then, researchers around the world have paid attention to extracellular vesicles, and exosome research in various fields has developed dramatically. However, an exosome is still something with a lipid bilayer, and there is no absolute indicator other than that it is a vesicle where tetraspanins such as CD9 and CD63 are detected on the surface. Therefore, to date, there are many studies where 'exosomes were extracted' from the size of vesicles that were detected in a certain size fraction by ultracentrifugation, or the size confirmed from vesicles that were treated with various commercial extraction kits followed by the detection of tetraspanin.
In our analysis, the amount of tetraspanin on the surface is not consistent, and the presence or absence of expression varies greatly according to cell type and state, as well as the type of drug. This should be kept in mind while using a kit for quantification, which utilizes antibody reaction against tetraspanins such as CD9 and CD8. In other words, you need to pay attention to the possibility of "only seeing what you want to see." We are conducting analyses using a simple kit (MagCaptureTM Exosome Isolation Kit PS) based on ultracentrifugation or adhesion to lipid as much as possible, so that surface markers do not become exosomes of a specific population.
2) Cautions in exosome administration experiments
A technique where extracted exosomes are labeled and administered to other cells or animals for analysis is also being used. Labeling using a fluorescence probe followed by analysis using exosome imaging12,13), and an exosome labeling method using iron oxide nanoparticles as a probe14,15) are also being used. However, it should be noted that administering a large amount of purified exosomes may lead to a result that is different from the original physiological function. There are also routes where a reverse reaction is seen when the amount is high. Therefore, confirming the concentration is an important element. However, the "what happens if the switch is pressed?" type of analytical studies, which are dosing experiments that have been commonly performed to date, are no doubt one established exosome study procedure16-21).
Co-culture studies to understand 'biological PINs'
There is a research methodology that clarifies the problems of exosome research that have been mentioned as points of caution to date. That is the co-culture of cells that are excreted without extraction and made to indirectly interact in an unmodified manner. With cell co-culture, the natural interaction of each cell can be observed. Additionally, the natural interaction between cells is a study in which the "biological PIN" is pressed between cells, and a phenomenon that is different from that of simply pressing a switch is also commonly observed. In other words, in order to decipher the "biological PIN," we would like to recommend co-culture studies, which are cell-cell interaction studies.
Co-culture studies became evident in the 1980s as intercellular communication studies and have been used in various research areas since then. The main motivation to use the co-culture system for research is to study the interaction between cells and microorganisms 22), and to develop new cell engineering methods using such interactions 23,24). In co-culture studies on exosomes, there are methods of studying the intracellular dynamics and uptake into cells by fluorescent labeling of CD63, CD81, CD9, etc., which are surface markers of exosomes. The advantage of these studies is that the natural actions can be observed through exosomes. However, the expression of surface markers such as CD63 varies depending on the cell type and condition, and some cells do not express some surface markers. In addition, there are still problems with optical properties, such as the accuracy of identifying labeled molecules in research using fluorescent labels. However, these methods are important research methods for studying cell-cell interactions performed i>in vivo. Exosome research using the co-culture system has been growing. For example, a report where an attempt was made to visualize the behavior of exosomes using a fusion protein of CD63, a protein located on the surface of exosomes, and GFP11); a report where it became evident that mesenchymal stroma stem cell (MSC)-derived exosomes that were taken up by breast cancer cells acquired MSC-derived cell capacity25); and MSC-derived CD90 was expressed in a co-culture25). For an experiment using fluorescent labeling, there is a report 25-27) confirming exosome exchange and migration. Even to date, experiments using the lower and bottom type of co-culture chambers have been an important model for culture studies where the cells are not mixed. A report has proved that vascular growth was suppressed by a co-culture of cardiomyocytes of diabetic model rat and mouse
myocardial intimal cells, and that exosomes and miR-320 in exosomes were involved27). Moreover, there is also a report that virus-derived miRNAs are transmitted to other cells via exosomes28). It has also been reported that the properties of cells change via exosomes when cells are cultured above and below the membrane of a Transwel® co-culture plate and do not come into direct contact with each other 29,30). There is a report on the use of the Transwell® co-culture plate in an in vitro BBB model 13). This is a model where three types of cells are cultivated, namely, cerebrovascular endothelial cells, pericytes, and astrocytes, thus simulating the blood-brain barrier (BBB). These models make good use of co-culture plates.
Co-culture study methods
The available co-culture systems are generally divided into two groups, contact direct co-culture and non-contact indirect co-culture, depending on the state of cell-cell adhesion. The method of mixing cells is direct co-culture, and the cells are in direct contact. Direct co-culture is generally performed effectively, but it is difficult to analyze individual cells. Moreover, it can only be analyzed as a cell population. It should be noted that not only the direct effects of contact but the effects of liquid factors are also involved. This makes it difficult to interpret the results.
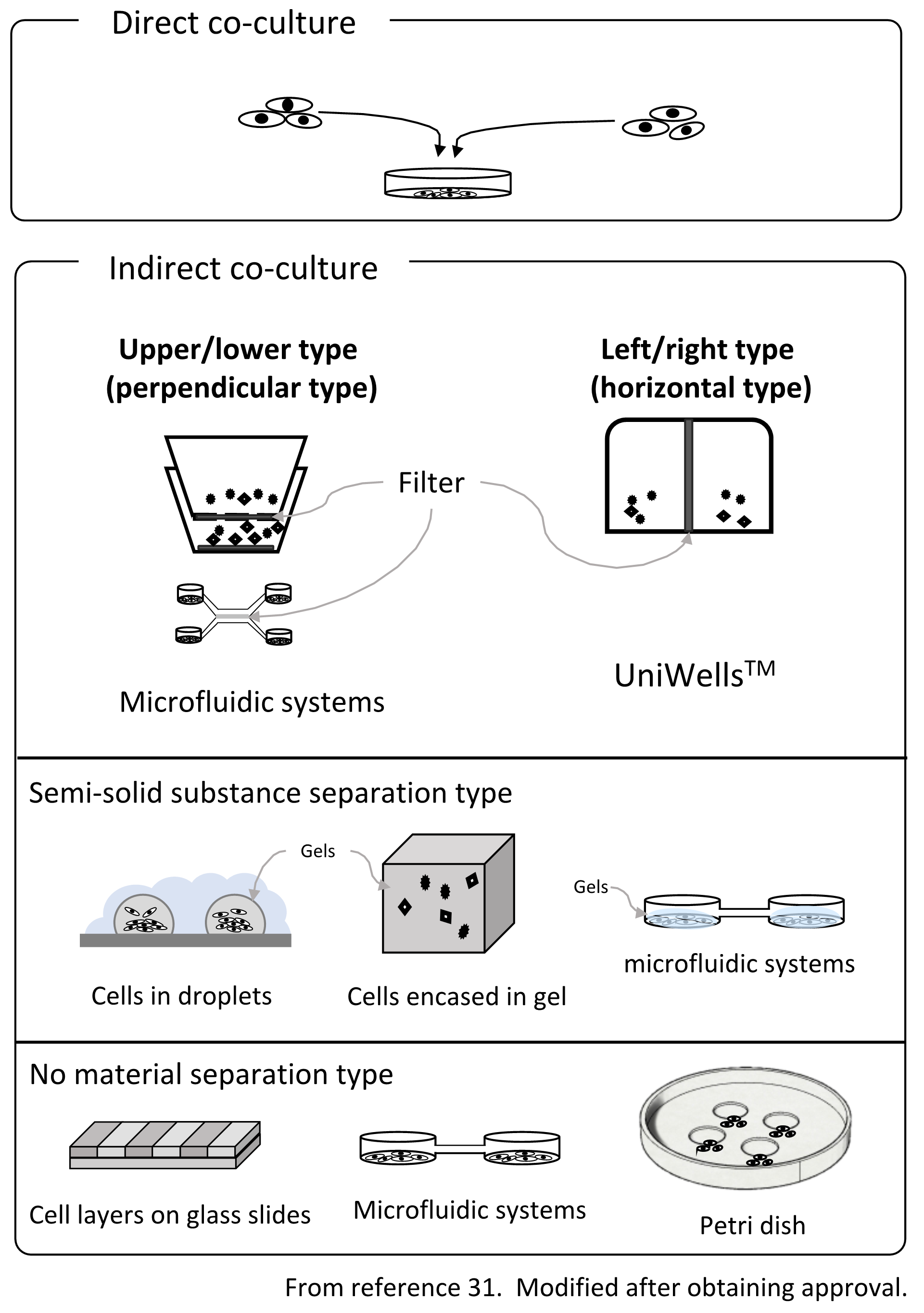
Fig. 2. Outline of the co-culture method
On the other hand, with indirect co-culture, cells are placed in separate environments, and cell-cell interactions occur via humoral factors. Since the cells can be observed after placing them in a state where they are not in direct physical contact using culture vessels, filters, gels, etc., the effects of indirect humoral factors can be clarified. Indirect co-culture techniques can be divided into three main methods31) (Figure 2.). (1) A type that separates with a filter, (2) a system type that uses semi-solids such as a gel to separate cells and exchange factors via the gel, and (3) a method where although complete separation is not possible, cells are cultivated in a colony using level differences, water drops, etc. In addition to these methods, the method of exchanging the culture solution32) is also used in research on bacteria. Boyden chamber33-35 is a co-culture chamber that uses a filter to connect the chambers vertically. Alternatively, the Transwell® co-culture plate with a modified Boyden chamber has been used as the standard. The Boyden chamber was developed by Stephan Boyden in 1962. Culture chambers are connected to the upper and lower chambers, while the bottom of the upper culture chamber is filtered. This is a method of co-culturing through a filter. Besides the name Transwell® co-culture plate, it is also often called 'Cell culture insert' as a general name. Initially, it was used to evaluate invasion, etc., but has come to be used as a standard method for many indirect co-cultures. However, it is difficult to obtain a fine image of the cells in the upper culture chamber due to the problem of the focal length of the microscope. In addition, the material of the bottom of the upper cell culture chamber is different from that of the lower chamber, which is a disadvantage. Cell behavior is often affected by the material on the bottom surface, and the effect of different materials on the bottom surface is significant. Also, since it is necessary to fabricate the chambers by integrating the filter, there are fewer types of filters.
In addition, even in terms of efficiency related to co-culture, the upper and lower type of co-culture chambers have a structure in which one bucket is put inside another due to its structural characteristics. For this reason, the volume ratio of the containers varies greatly, such as 1:3, and the humoral factors secreted by the cells are diluted. In addition, since cells in the upper chamber are located on the filter, if the cell density increases, the holes in the filter are blocked, and the co-culture effect decreases with time.
Surprisingly, there was no laterally connected filter-separation type of chambers. Recently, however, there have been reports of lateral containers for bacterial research32), but not for observation purposes. We have developed a horizontal co-culture container that can be observed with a microscope, using a novel cell culture chamber with a lateral connection (UniWellsTM: sold by Fuji Film Wako Pure Chemical Industries, Ltd.).
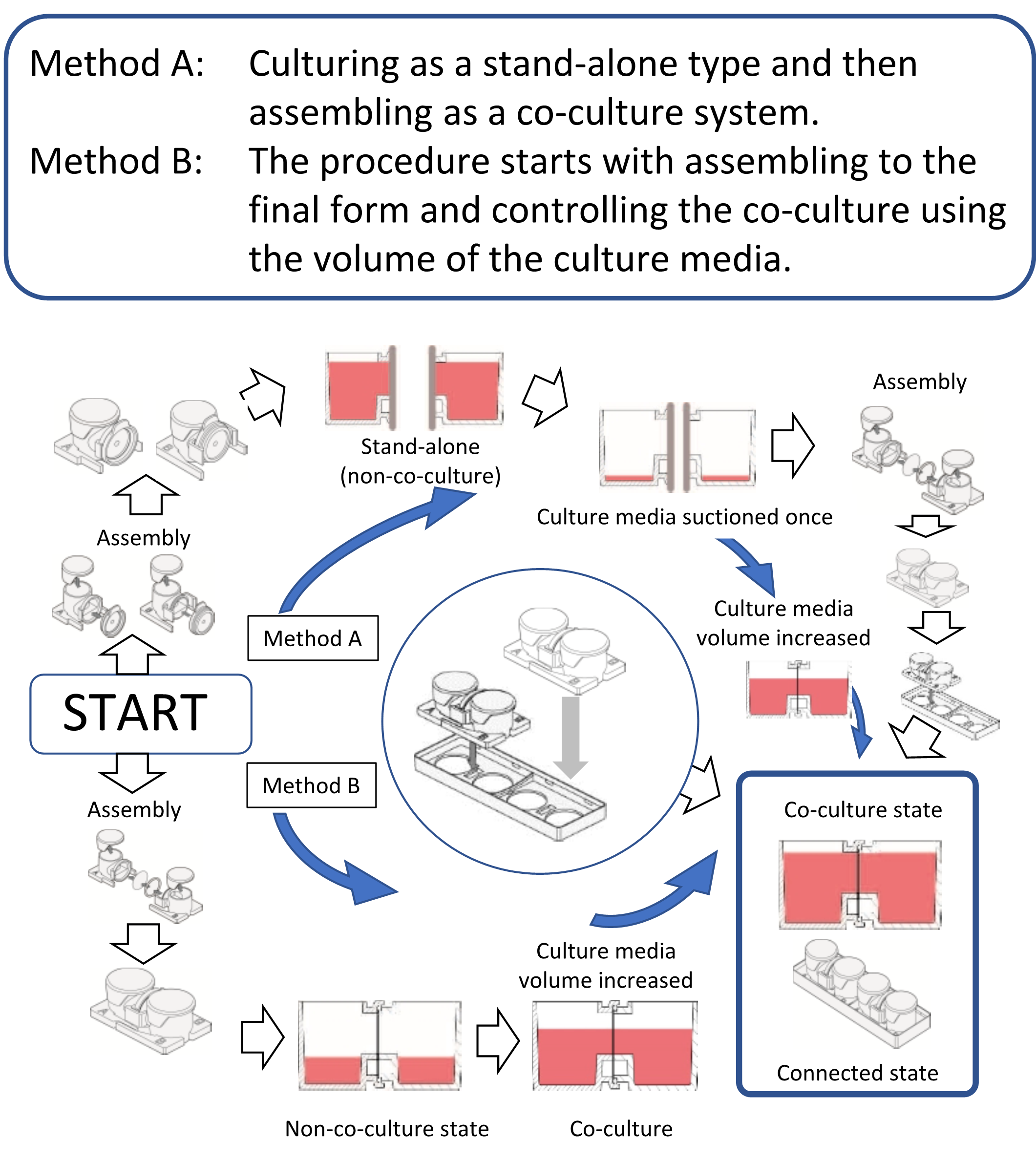
Fig. 3 Usage of co-culture chambers
Since we have developed a co-culture chamber31), this article introduces a culture chamber in which any commercially available filter can be placed between the chambers. Indeed, two containers can be combined at any time. It is also possible to easily control the co-cultivation state by performing the culturing in separate environments and then bringing it into the co-culture system and also controlling the volume of the culture medium (Figure 3). The greatest merit of this horizontal co-culture vessel is that the positional relationship between the cells and the filter is distant. Moreover, since the volume ratio of the container is 1:1, the co-culture effect shows maximum efficiency, and both cells can be observed simultaneously from the lower surface. There are two methods of use. One is the method of co-culturing after culturing in single chambers by connecting the two chambers. The other is a method in which two containers are connected and assembled from the onset, and the co-culture state is controlled using the height to the connection surface. Assembly is easy, and researchers can choose either of the two methods to use. Co-culture in 3D culture is also possible by performing a 3D culture using gel or the like. Cell culture insert type chambers connect in the vertical direction, so both cells cannot be observed simultaneously with a microscope. However, the co-culture chamber we developed is connected horizontally, and so both cells can be observed simultaneously. Regardless of whichever system is used, co-culture systems are effective for research where cell-cell interactions are observed and for cell engineering techniques that utilize cell interactions. The co-culture system can provide various methods for cell engineering and is also considered to be very effective for drug discovery24).
Future of exosome research
The involvement of exosomes has become clear not only in the fields of cancer, immunological diseases, neurological diseases, and regenerative medicine, where various studies have been developed but also in diseases for which the cause and mechanism are unknown. Research on exosomes and diseases will likely reveal the relationship between diseases and exosome inclusions, although there is some caution regarding the fact that exosome extraction methods are not standardized. However, knowledge is still lacking regarding the physiological functions of exosomes and the basic mechanisms of their secretion and uptake. To achieve innovative research, exosome research using a co-culture system is important. It is hoped that the "biological PIN" via exosomes will be elucidated in the future as a result of a lot of pioneering research using the co-culture system.
Future of exosome research
The involvement of exosomes has become clear not only in the fields of cancer, immunological diseases, neurological diseases, and regenerative medicine, where various studies have been developed but also in diseases for which the cause and mechanism are unknown. Research on exosomes and diseases will likely reveal the relationship between diseases and exosome inclusions, although there is some caution regarding the fact that exosome extraction methods are not standardized. However, knowledge is still lacking regarding the physiological functions of exosomes and the basic mechanisms of their secretion and uptake. To achieve innovative research, exosome research using a co-culture system is important. It is hoped that the "biological PIN" via exosomes will be elucidated in the future as a result of a lot of pioneering research using the co-culture system.
References
- Taylor, D. D. and Gercel-Taylor, C. : Oncol., 110, 13 (2008).
- Murakami, Y. et al. : PLoS One, 7, e48366 (2012).
- Moon, P. G. et al. : Proteomics, 11, 2459 (2011).
- Hoshino, A. et al. : Nature, 527, 329 (2015).
- Kosaka, N. et al. : Clin. Invest., 126, 1163 (2016).
- Valadi, H. et al. : Cell Biol., 9, 654 (2007).
- Ratajczak, J. et al. : Leukemia, 20, 847 (2006).
- Chargaff, E. and West, R. : Biol. Chem., 166, 189 (1946).
- Wolf, P. : J. Haematol., 13, 269 (1967).
- Johnstone, R. M. et al. : Biol. Chem., 262, 9412 (1987).
- Yanez-Mo, M. : Extracell. Vesicles, 4, 27066 (2015).
- Smyth, T. et al. : Control. Release, 199, 145 (2015).
- Wiklander, O. P. et al. : Extracell. Vesicles, 4, 26316 (2015).
- Busato, A. et al. : J. Nanomedicine, 11, 2481 (2016).
- Hu, L. et al. : Reson. Med., 74(1), 266 (2014).
- Chen, Y. et al. : Oncogene, 36, 4692 (2017).
- Yuyama, K. et al. : Neurochem., 105, 217 (2008).
- Rappa, G. et al. : Cancer, 12, 62 (2013).
- Chowdhury, R. et al. : Oncotarget, 6, 715 (2015).
- Saeed-Zidane, M. et al. : PLoS One, 12, e0187569 (2017).
- Ekstrom, K. et al. : Extracell. Vesicles, 1 (2012). doi: 10.3402/jev.v1i0.18389
- Cottet, S. et al. : Biol. Chem., 277, 33978 (2002).
- Tanouchi, Y. et al. : Opin. Biotechnol., 23, 791 (2012).
- Moraes, C. et al. : Biomed. Eng., 40, 1211 (2012).
- Yang, Y. et al. : J. Oncol., 47, 244 (2015).
- Hergenreider, E. et al. : Cell Biol., 14, 249 (2012).
- Wang, X. et al. : Mol. Cell. Cardiol., 74, 139 (2014).
- Shin, Y. et al. : Protoc., 7, 1247 (2012).
- Su, M. J. et al. : Rep., 6, 30110 (2016).
- Li, Y. et al. : Stem Cell Res, Ther., 8, 198 (2017).
- Shimasaki, T. et al. : Pharm. Bull., 41, 1311 (2018).
- Moutinho, T. J., Jr. et al. : PLoS One, 12, e0182163 (2017).
- Boyden, S. : Exp. Med., 115, 453 (1962).
- Thomsen, R. and Lade Nielsen, A. : Glia, 59, 1782 (2011).
- Albini, A. et al. : Cancer Res., 47, 3239 (1987).




