ISOSPIN Plant DNA
- Manufacturer :
- Nippon Gene Co., Ltd.
- Storage Condition :
- Keep at RT.
- GHS :
-
- Structural Formula
- Label
- Packing
|
Comparison
|
Product Number
|
Package Size
|
Price
|
Availability
|
Certificate of Analysis
|
Purchase |
|---|---|---|---|---|---|---|
|
|
|
50Tests
|
|
Out of Stock |
|
※Check availability in the US with the distributor.
Document
- Package Insert
-
- Spectral Data
-
- Certificate of Analysis
-
- Calibration Certificate
-
- Analytical Charts
-
Kit component
For 50 preps
| Prewash Buffer | 30 mL x 1 |
|---|---|
| PE1 Buffer | 22.5 mL x 1 |
| PE2 Buffer | 2.5 mL x 1 |
| PB Buffer | 30 mL x 1 |
| PW1 Buffer | 40 mL x 1 |
| PW2 Buffer | 45 mL x 1 |
| RNase A (100 mg/mL) | 250 μL x 1 |
| Elution Buffer | 3 mL x 1 |
| Spin Column | 50 columns x 1 |
Description
ISOSPIN Plant DNA is a kit for the extraction and purification of DNA from plant leaves.
This kit employs a Centrifuge-based method that effectively removes impurities, enabling the extraction of high-purity DNA even from plant samples containing large amounts of polyphenols and viscous substances, which have traditionally been difficult to process.
Product features
- Efficient DNA extraction from highly viscous Rosaceae plant tissues
- No need for phenol or chloroform
- Proprietary spin column ensures excellent usability
- RNase A included
Protocol
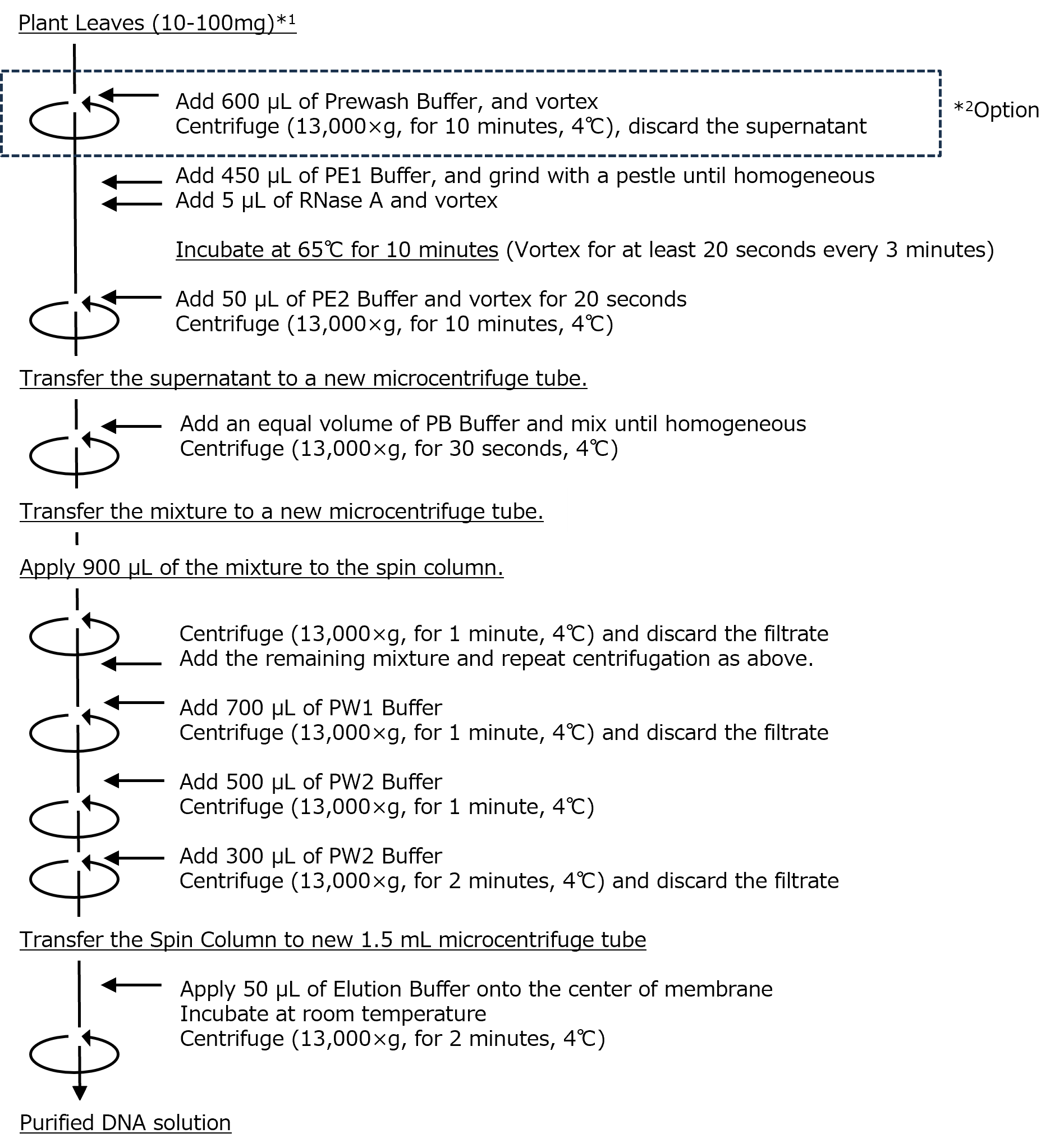
Recommended sample volume
- Samples for which optional steps can be omitted:
Spinach (50 mg), Arabidopsis thaliana (20 mg), Cabbage (100 mg), Rice (50 mg), Chrysanthemum (50 mg), Pine (20 mg) - Samples for which optional steps are required:
Strawberry (20 mg), Rose (50 mg), Cherry blossom (50 mg), Kiwifruit (20 mg), Cedar needles (10 mg)
Data
DNA extraction from spinach leaves
| Sample | PreWash* | Amount | ng/μL | A260/A280 | A260/A230 |
|---|---|---|---|---|---|
| Spinach | - | 56.4 mg | 44.4 | 1.82 | 2.97 |
| 56.8 mg | 35.2 | 1.84 | 3.07 | ||
| 57.5 mg | 39.6 | 1.84 | 3.05 | ||
| 58.8 mg | 36.3 | 1.83 | 2.96 |
Electrophoresis
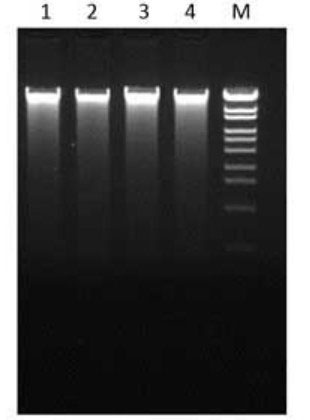
Run 1/10 volume of DNA extraction solution (5 μL).
1:56.4 mg of spinach sample
2:56.8 mg of spinach sample
3:57.5 mg of spinach sample
4:58.8 mg of spinach sample
M:OneSTEP Marker 6
In 1% Agarose S
DNA extraction from cedar needles
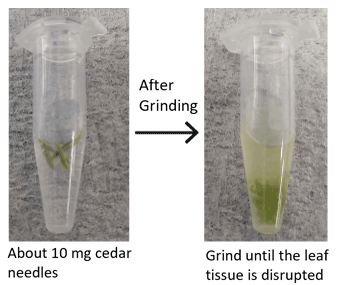
DNA was extracted from cedar needles following the ISOSPIN Plant DNA kit manual.
To obtain high-purity DNA:
- Add Prewash Buffer and grind the leaf tissue thoroughly with a pestle until it is completely disrupted (see photo).
- After the Prewash step, carefully pipette out and discard the supernatant, which is highly viscous and stringy.
- Continue with the protocol (starting from the PE2 Buffer addition and centrifugation step).When collecting the supernatant, transfer it carefully to a new microcentrifuge tube, avoiding the pellet and any precipitates on the surface as much as possible. For cedar samples, collect approximately 350 µL of supernatant.
| sample | PreWash | Amount | ng/μL | A260/A280 | A260/A230 |
|---|---|---|---|---|---|
| Cedar needles | 12.1 mg | 26.3 | 1.81 | 2.62 | |
| 14.6 mg | 25.7 | 1.82 | 2.20 | ||
| 13.4 mg | 29.5 | 1.79 | 2.80 | ||
| 13.7 mg | 30.3 | 1.82 | 2.63 |

Run 1/10 volume of DNA extraction solution (5 μL)
1:12.1 mg of cedar needles sample
2:14.6 mg of cedar needles sample
3:13.4 mg of cedar needles sample
4:30.3 mg of cedar needles sample
M:OneSTEP Marker 6
In 1% Agarose S
FAQ
- Can I extract DNA from samples other than plant leaves?
- Yes. We have successfully extracted DNA from husk, chestnut (cotyledon), sweet potato, and beech mushroom (stem).
- How should I perform re-purification of a DNA solution that was crudely extracted from a plant sample using this kit?
- Please follow the protocol below:
Re-purification Protocol
Prepare the DNA solution (≤100 µL)
↓←450 μL of PE1 Buffer
↓[←5 μL of RNase A (100mg/mL), mix well, and incubate at room temperature for 10 minutes] *
↓←50 μL of PE2 Buffer and vortex for at least 20 seconds
Centrifuge (13,000×g, for 10 minutes, 4℃)
↓
Transfer the supernatant to a new microcentrifuge tube
↓←Add an equal volume of PB Buffer and mix by inversion until homogeneous
↓
Centrifuge (13,000×g, for 30 seconds, 4℃)
↓
Transfer the mixture to a new microcentrifuge tube
↓
Apply 900 µL of the mixture to the spin column
(Continue following the standard protocol described in the manual)
*If the DNA solution has already been treated with RNase, the RNase A treatment step (enzyme addition and 10-minute incubation at room temperature) can be omitted.
- Can I extract DNA from mushrooms?
- Yes. We have successfully extracted DNA from Flammulina velutipes (enoki mushroom), Hypsizygus marmoreus (beech mushroom), and Pholiota microspora (nameko mushroom).
Since mushrooms exhibit strong DNA-degrading activity, we recommend adding 20 µL of Proteinase K (20 mg/mL) after the RNase A treatment step.
Overview / Applications
Property
Manufacturer Information
Alias
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.







