Ethachinmate
- Manufacturer :
- Nippon Gene Co., Ltd.
- Storage Condition :
- Keep at 2-10 degrees C. (RT)
- Structural Formula
- Label
- Packing
|
Comparison
|
Product Number
|
Package Size
|
Price
|
Availability
|
Certificate of Analysis
|
Purchase |
|---|---|---|---|---|---|---|
|
|
|
0.02mL
|
|
In stock in Japan |
|
|
|
|
|
0.2mL
|
|
In stock in Japan |
|
※Check availability in the US with the distributor.
Document
- Package Insert
-
- Spectral Data
-
- Certificate of Analysis
-
- Calibration Certificate
-
- Analytical Charts
-
OverView
"Ethachinmate" is a high–molecular-weight carrier used for the precipitation of nucleic acids (DNA and RNA) with ethanol or isopropanol. It enables the efficient recovery of trace amounts of nucleic acids. It has been tested to be DNase-free and RNase-free.
Feature
・Recovery of trace nucleic acids
Enables quantitative recovery of DNA (>100 base pairs) and RNA (>120 bases) at concentrations above 20 ng/mL.
・Rapid alcohol precipitation
No incubation at –20 °C or –80 °C is required. Centrifugation can be performed immediately after the addition of alcohol.
・No inhibition of enzymatic reactions
Ethachinmate itself forms a visible pellet upon alcohol precipitation, reducing the risk of losing valuable nucleic acids, even in trace amounts, during washing.
・Visible precipitation
Once the ethanol is added, Ethachinmate itself forms a visible pellet. The risk of losing the pellet by washing is therefore reduced.
・Tested DNase and RNase-free
It has been confirmed to be DNase and RNase-free.
Protocol
DNA or RNA solution (100 µl)
↓←3.3 µl, 3 mol/l Sodium Acetate(Product Component) *1
↓←1 µl, Ethachinmate *2
↓vortex *3
↓←200 - 250 µl, Ethanol
↓vortex *3
↓12 K x g for 5min at room temperature *4
pellet *5
- *1 Final salt concentration must be more than 0.1 mol/l.
- *2 Add 1 µl of Ethachinmate per 100 µl of DNA solution. If the amount of DNA solution is less than 100 µl, add 1 µl of Ethachinmate. If the amount of DNA solution is more than 300 µl, 3 µl of Ethachinmate is the correct amount to be added. Once Ethachinmate is added into DNA solution, there is no need to add Etachinmate again if performing more centrifugations. Adding to much Ethachinmate may make the solution viscous, causing the following processes to be difficult.
- *3 Vortexing improves the efficiency to recover subtle DNA.
- *4 Cooling is not necessary.
- *5 Pellet is visible. Pellet may then be dissolved in buffer and used as a template for enzyme reactions. Wash with 70 % ethanol, if needed.
RNA extraction protocol using ISOGEN with Ethachinmate
When total RNA is extracted using ISOGEN, the RNA pellet ma sometimes be difficult to see.
By adding Ethachinmate, the pellet is easier to see.
Sample
↓←ISOGEN
↓Homogenize and incubate at room temperature for 5min.
↓←Chloroform
↓Shake vigorously and incubate at room temperature for 3min.
↓Centrifuge for 15 min to separate the phases
↓Transfer the aqueous phase to a new tube
↓←1-2 µl of Ethachinmate *1
↓vortex
↓←Isopropanol.
↓Mix by gentle inversion and incubate at room temperature for 10 min.
↓Centrifuge for 10 min.
↓Remove the supernatant.
Pellet
↓←70 % Ethanol
↓vortex
↓Centrifuge for 5 min.
↓Remove the supernatant
Pellet
↓Air-dry the pellet.
↓←RNase-free water or TE buffer (pH 8.0) for dissolving the pellet.
Total RNA Solution
Data
Recovery of Trace Amounts of Nucleic Acid
λ/HindIII fragments (10 ng in 500 µL) were precipitated using 1 mL of ethanol in the presence of 0.1 mol/L sodium acetate and analyzed by agarose gel electrophoresis.

Lane 1 : λ/HindIII (Contorol; no precipitation treatment)
Lane 2 : λ/HindIII+Ethachinmate 3 µl (immediate centrifugation)
Lane 3 : λ/HindIII Incubation at -20 °C overnight
Lane 4 : λ/HindIII Incubation at -80 °C for 20 min.
Result
Under the condition of 0.1 mol/L sodium acetate, adding Ethachinmate allows for quantitative recovery of trace amounts of DNA without the need for low-temperature incubation.
Low-Molecular-Weight RNA Ethanol Precipitation
Synthetic 21mer RNA samples (10 ng, 20 ng, 40 ng, and 80 ng) were subjected to ethanol precipitation under the following conditions:
| Sample | Synthesized 21 mer RNA (10 ng,20 ng,40 ng,80 ng) |
|---|---|
| Ethanol precipitation | Each 100 µL RNA solution was added 3M Sodium acetate 3.3 µL and Ethachinmate 1 µL. All samples were immediately precipitated after adding Ethanol. |
| Electrophoresis | 15 % polyacylamide gel |

| RNA Volume | ||
|---|---|---|
| Lane 1 | Before Ethanol Precipitaion |
10ng |
| Lane 2 | 20ng | |
| Lane 3 | 40ng | |
| Lane 4 | 80ng | |
| Lane 5 | Without Ethachinmate |
10ng |
| Lane 6 | 20ng | |
| Lane 7 | 40ng | |
| Lane 8 | 80ng | |
| Lane 9 | With Ethachinmate |
10ng |
| Lane 10 | 20ng | |
| Lane 11 | 40ng | |
| Lane 12 | 80ng | |
Affect for enzymes
Data1. Restriction Enzyme Digestion and T4 DNA Ligase
λ DNA was subjected to digestion, ligation, and re-digestion using restriction enzymes (Hae III, Hind III) and T4 DNA Ligase, in the presence or absence of Ethachinmate (1 µL per 10 µL reaction mixture).
| Restriction Enzyme | Hae Ⅲ | Hind Ⅲ | Lane 1: Cut Lane 2: Cut and ligase Lane 3: Cut and ligase and re-cut |
||
|---|---|---|---|---|---|
| Etachinmate | - | + | - | + | |
| Result |  |
 |
 |
 |
|
Result
Ethachinmate did not affect the anzyme activity of restriction ligation efficiency.
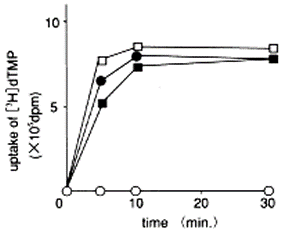
Data2. Reverse Transcriptase
In the presence of Ethachinmate, [3H]dTMP was incorporated into the poly(rA)·(dT)12-8 template using AMV Reverse Transcriptase (RTase).
Reverse Transcription Reaction
50 µL [50 mmol/l Tris-HCl(pH8.3), 6 mmol/l MgCl2, 40 mmol/l KCl, 0.5 mmol/l [3H]dTTP, 0.4 mmol/l poly(rA)・(dT)12-8, 5 units AMV RTase]
Temperature: 42 ℃
| RTase | Ethachinmate | |
|---|---|---|
| - | 10 µL |  |
| + | 0 µL |  |
| + | 5 µL | 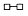 |
| + | 10 µL | 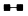 |
Result
Ethachinmate did not affect the activity of Reverse Transcriptase.
Data3. Taq DNA Polymerase
In the presence of Ethachinmate, a DNA (about 600 bp) was amplified using Taq DNA Polymerase.
PCR Reaction
25 µL [TAPS-HCl, pH9.3, 50 mmol/l KCl, 2 mmol/l MgCl2, 1 mmol/l 2-Mercaptoethanol, 0.01 % gelatin, 200 µmol/l dNTPs, 0.2 M primer, 1.25 units Taq DNA Polymerase]

Lane 1 : Ethachinmate 0 µl
Lane 2 : Ethachinmate 0.2 µl
Lane 3 : Ethachinmate 0.5 µl
Lane 4 : Ethachinmate 1 µl
Lane 5 : Ethachinmate 3 µl
Lane 6 : Ethachinmate 5 µl
Result
Ethachinmate did not affect the activity of Taq DNA Polymerase.
FAQ
- Is it OK to use for RNA precipitation?
- It is OK. It can be used to recover RNA.
- Are there any DNase or RNase contaminants?
- No. Ethachinmate has been tested and confirmed to be DNase-free and RNase-free.
- Which length and concentration of DNA and RNA are recovered?
- It is possible to recover almost all of the DNA (> 100 base pairs) and RNA (> 120 bases). If the concentration of nucleotide is less than 20 ng/ml, efficiency may lower.
- Does it have any effect on quantitation by absorbance at 260 nm?
- No. It does not affect quantitation by A260 measurement.
- My DNA solution is less than 100 µl. How much Ethachinmate should I use?
- Add 1 µl of Ethachinmate. Making the DNA pellet visible requires over 1 µl of Ethachinmate. Calculate the amount of 3 mol/l Sodium Acetate to add into your DNA solution based on its volume. (e. g., for 50 µl of DNA solution, add 1 µl of Ethachinmate and 1.7 µl of 3 mol/l Sodium Acetate).
- How much Ethachinmate should I use if the DNA solution volume is more than 300 µL?
- 3 µL of Ethachinmate is sufficient even for larger volumes. Do not add more than this. Adjust sodium acetate proportionally (e.g., for 600 µL DNA solution, add 3 µL Ethachinmate and 19.8 µL of 3 M sodium acetate).
- I want to perform ethanol precipitation two or more times. Should I add Ethachinmate for each precipitation?
- No. Once you add Ethachinmate into DNA solution there is no need to add Ethachinmate again. Multiple additions of Ethachinmate may make the solution viscous, causing subsequent processes to become more difficult.
- Does freezing affect quality of Ethachinmate?
- No, it doesn't affect the quality.
- Does autoclave freeze affect quality of Ethachinmate?
- No, it doesn't affect the quality.
- Does phenol / chloroform treatment of DNA solution containing Ethachinmate bring any effect on the quality of Ethachinmate?
- No, it doesn't affect the quality.
- Does using Ethachinmate affect band pattern of gel electrophoresis?
- No. However, depending on the electrophoresis conditions, DNA bands larger than several tens of kbp may appear broad.
- Does Ethachinmate affect digestion using restriction enzyme?
- No, it doesn't affect digestion.
- Does Ethachinmate affect ligation using T4 DNA ligase?
- No, it doesn't affect ligation.
- Does Ethachinmate affect cDNA synthesis using AMV reverse transcriptase?
- No, it doesn't affect cDNA synthesis.
- Does Ethachinmate affect PCR using Taq DNA polymerase?
- No, it doesn't affect PCR.
- Does Ethachinmate affect reaction of Klenow Fragment?
- No, it doesn't affect.
- Does Ethachinmate affect transformation of E. coli?
- No. It does not affect electroporation either. However, please note that when Ethachinmate, PEG6000, or polyamines are present in a ligation product solution to be introduced into E. coli, the transformation efficiency may decrease.
- Does Ethachinmate affect in vitro Packaging?
- It slightly decreases the efficiency of lambda packaging.
- Does Ethachinmate affect transfection efficiency?
- When siRNA prepared using Ethachinmate was transfected with a lipofection reagent, a decrease in efficiency was observed. Although other transfection methods have not been tested, Ethachinmate may reduce transfection efficiency.
- Does ethanol precipitation with Ethachinmate precipitate mono nucleotide?
- Experiments using 8 and 17 base nucleotides did not show significant differences between the presence or the absence of Ethachinmate.
- Does Ethachinmate get denatured in hybridization solution containing formamide?
- No. It is not become denatured.
- Does Ethachinmate affect blotting?
- No, it doesn't affect blotting
- Does Ethachinmate affect sequencing?
- It does not affect cycle sequencing with ABI Prism® 377, or sequencing by dideoxy method.
- Does the addition of Ethachinmate make the pellet more likely to detach from the tube?
- It depends on the type of material of the tube. (e. g., "Safe-Lock tube" from Eppendorf tends to show this.)
- Can nucleic acids containing Etachinmate be used for Agilent Bioanalyzer analysis?
- When we analyzed RNA samples that contained an estimated 0.1 %–0.5 % carryover of Ethachinmate, no particular problems were observed. (This observation was made using the Agilent RNA 6000 Pico Kit; the effects on other Bioanalyzer kits have not been confirmed.)
Overview / Applications
| Outline | This product is for research use only. Do not administer it to human. Ethachinmate replaces tRNA used in ethanol precipitation to concentratenucleic acids. When a solution containing nucleic acids is diluted, a co-precipitantis often used for the recovery of the nucleic acids.Ethachinmate, a neutral polyacrylamide polymer solution, is a veryefficient carrier for precipitation of a very small quantity of nucleicacids with ethanol. Advantages:1. Allows for quantitative recovery of DNA at the concentration of20 ng/mL or more (100 bps) and RNA (120 bps).2. Incubation at -20 degrees C or -80 degrees C is not required.3. Ethachinmate does not inhibit any enzymatic reactions.4. Ethachinmate forms pellets when ethanol is added; thus, even smallquantities of DNA are visible. |
|---|
Property
Manufacturer Information
Alias
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.



