Protein phosphorylation analysis using Phos-tag-based techniques
This article was written by Emiko Kinoshita-Kikuta, Eiji Kinoshita, Tohru Koike Department of Functional Molecular Science, Institute of Biomedical and Health Sciences, Hiroshima University, Kasumi 1-2-3, Hiroshima, Japan.
Introduction
Proteins responsible for vital cellular functions use kinase-induced phosphorylation and phosphatase-induced dephosphorylation for their activation and deactivation, respectively. Wide varieties of diseases are caused by dysregulation of kinase and phosphatase activity. Various kinase abnormalities have been reported in cancer and neurodegenerative disorders. Therefore, the analysis of protein phosphorylation is a critical area of research that can lead to the identification of the cause for various diseases and can aid in the development of kinase inhibitors (molecular-targeted agents).
To date, we have developed techniques for protein phosphorylation analysis using Phos-tag, a functional molecule that specifically captures a phosphate group (-PO32-). This article introduces three Phos-tag-based techniques that are used by life science researchers.
Analysis of phosphorylated proteins using Phos-tag SDS-PAGE
Phos-tag SDS-PAGE is a phosphate-affinity electrophoresis technique that separates phosphorylated and non-phosphorylated proteins using conventional SDS-PAGE procedures.1, 2) This technique uses a separation gel in which acrylamide is co-polymerized with Phos-tag Acrylamide that acts as a phosphate-trapping molecule. The reagents, protein sample preparation, and operation of electrophoresis are similar to those for the conventional SDS-PAGE procedures. As phosphorylated proteins exhibit reversible binding to Phos-tag immobilized in the gel during electrophoresis, they migrate more slowly than do non-phosphorylated proteins, resulting in detectable differences in the mobilities of phosphorylated proteins (Figure 1).
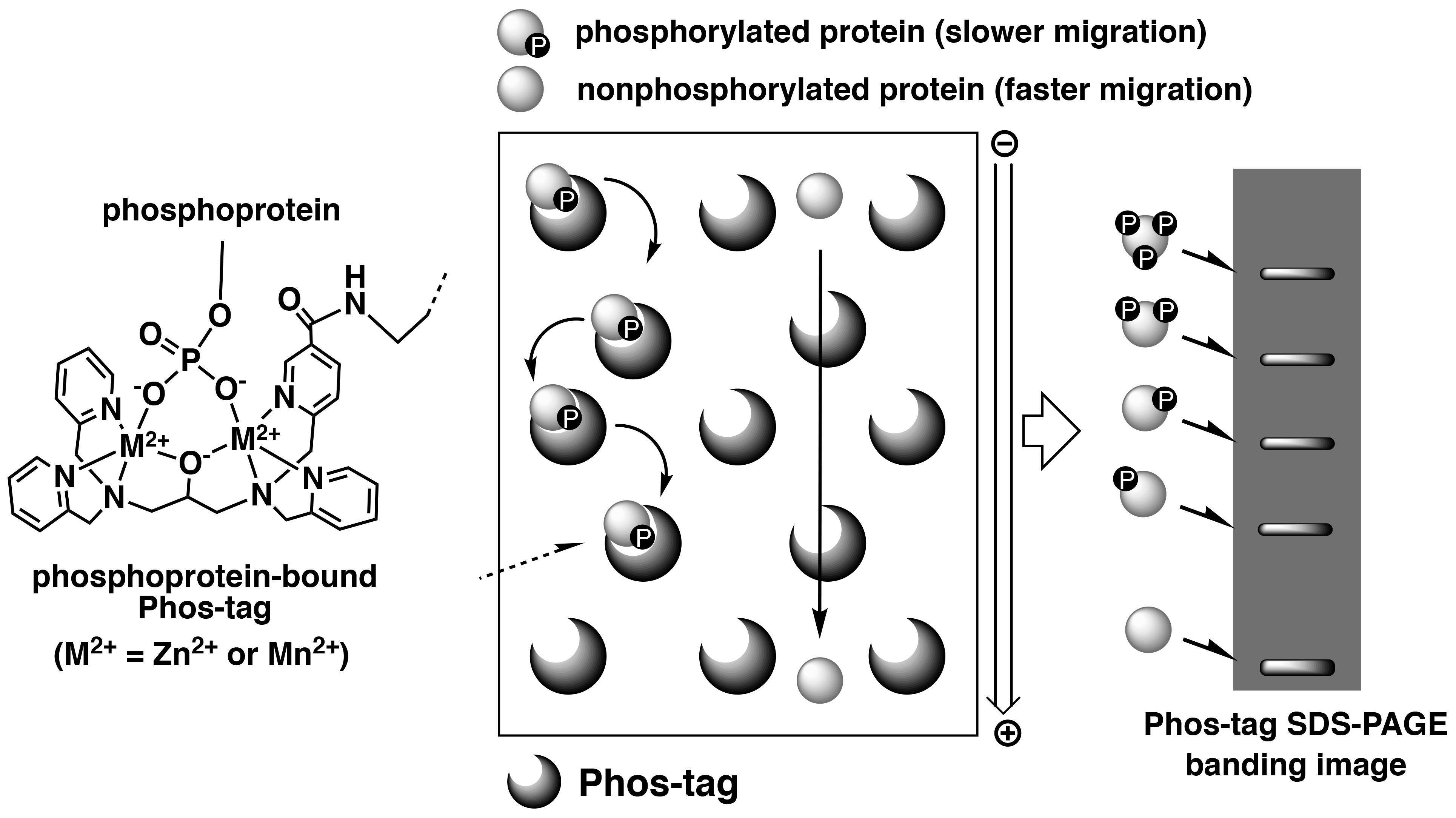
Figure 1.Overview of Phos-tag SDS-PAGE
This electrophoresis technique permits separation of the phosphorylated proteins that contain an identical number of phosphate groups, but at different locations within the molecule resulting in different migration bands. Hence, diverse phosphorylated forms of a single protein can be detected as multiple migration bands on the gel. Furthermore, Phos-tag SDS-PAGE enables analysis of not only the Ser-, Thr- and Tyr-phosphorylated proteins, but also unstable His- and Asp-phosphorylated proteins that are involved in a two-component signaling system.3) This technique is therefore appropriate for quantitative and qualitative analyses of proteins with different phosphorylation states. Owing to the commercial availability of the Phos-tag SDS-PAGE precast gels (the SuperSep Phos-tag series), this technique can be easily applied to multiple research studies.4) The banding images of phosphorylated proteins are available in the Phos-tag SDS-PAGE data collection.
Recently, we analyzed different phosphorylated forms of MEK1 in cancer using Phos-tag SDS-PAGE.5) Characteristic mutations of the MEK1-coding gene have been identified in certain sporadic cancers and in MEK-inhibitor-resistant cancer cells. Phos-tag SDS-PAGE revealed that the introduction of mutations can produce constitutively active MEK1 species containing phosphorylated Ser-218 and Ser-222 residues (Figure 2) and induces constitutive activity of the kinase. Phos-tag-based phosphorylation profiling of MEK1 can therefore provide clinical insights into the characteristics of individual mutations in the MEK1-coding gene.
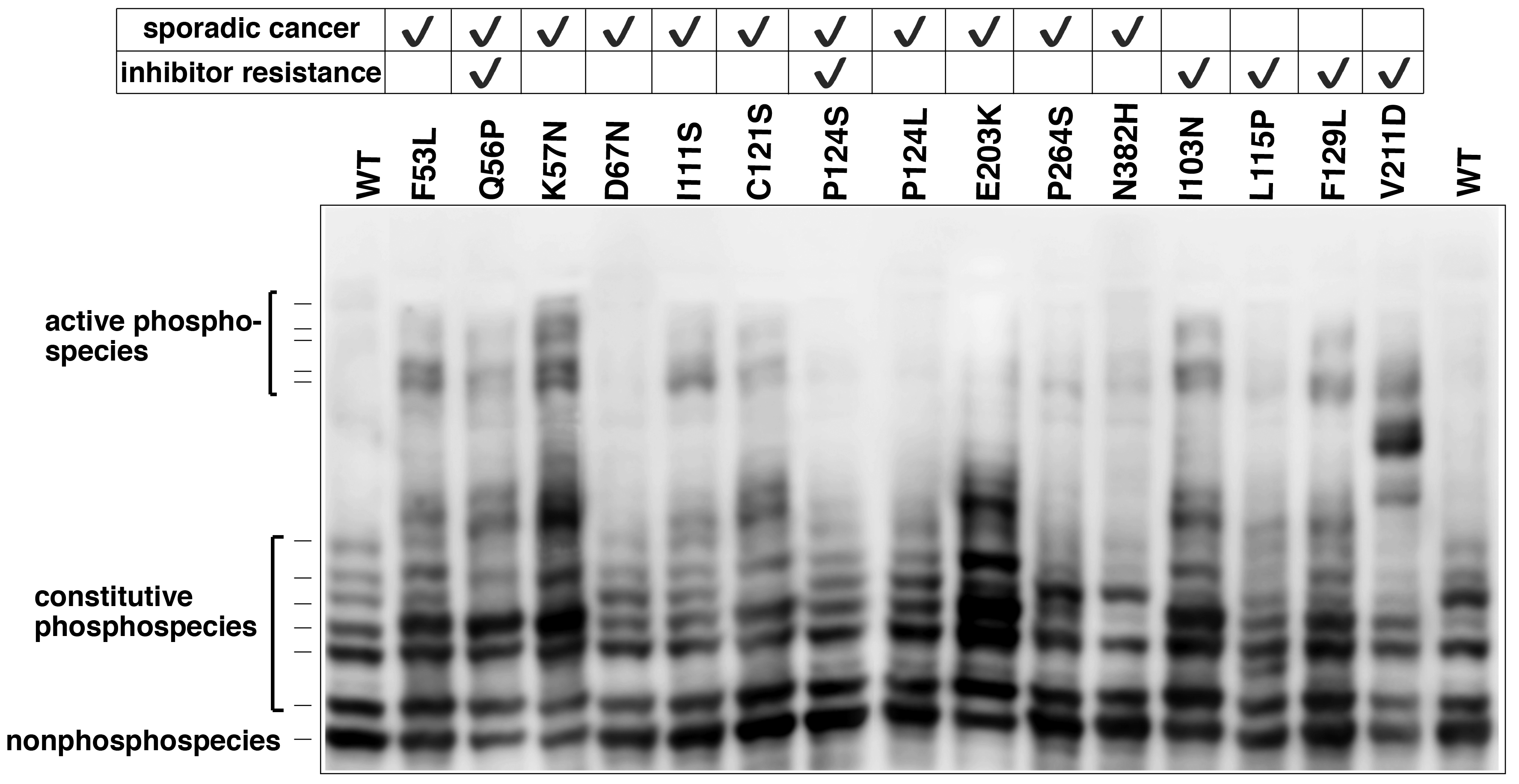
Figure 2. MEK1 mutations and increased MEK1 phosphorylation species in cancer (reprint from reference 5) The phosphorylation state of MEK1 mutants reported in lung cancer, ovarian cancer, and anti-cancer resistant cell lines was analyzed using Phos-tag SDS-PAGE.
Analysis of phosphorylated proteins using Phos-tag Biotin
Phos-tag Biotin can be used as a molecular tool in applications such as western blotting or microarray-based methods for comprehensive detection of phosphorylated proteins. In this technique, a complex is formed between Phos-tag Biotin and a horseradish peroxidase (HRP)-bound streptavidin tetramer in a 4:1 ratio, followed by probing of the blot membrane or microarray. Phosphorylated proteins that are specifically bound to Phos-tag Biotin are detected by the reaction between the chemiluminescence (ECL) substrate and HRP as enhanced ECL signals (Figure 3).1)
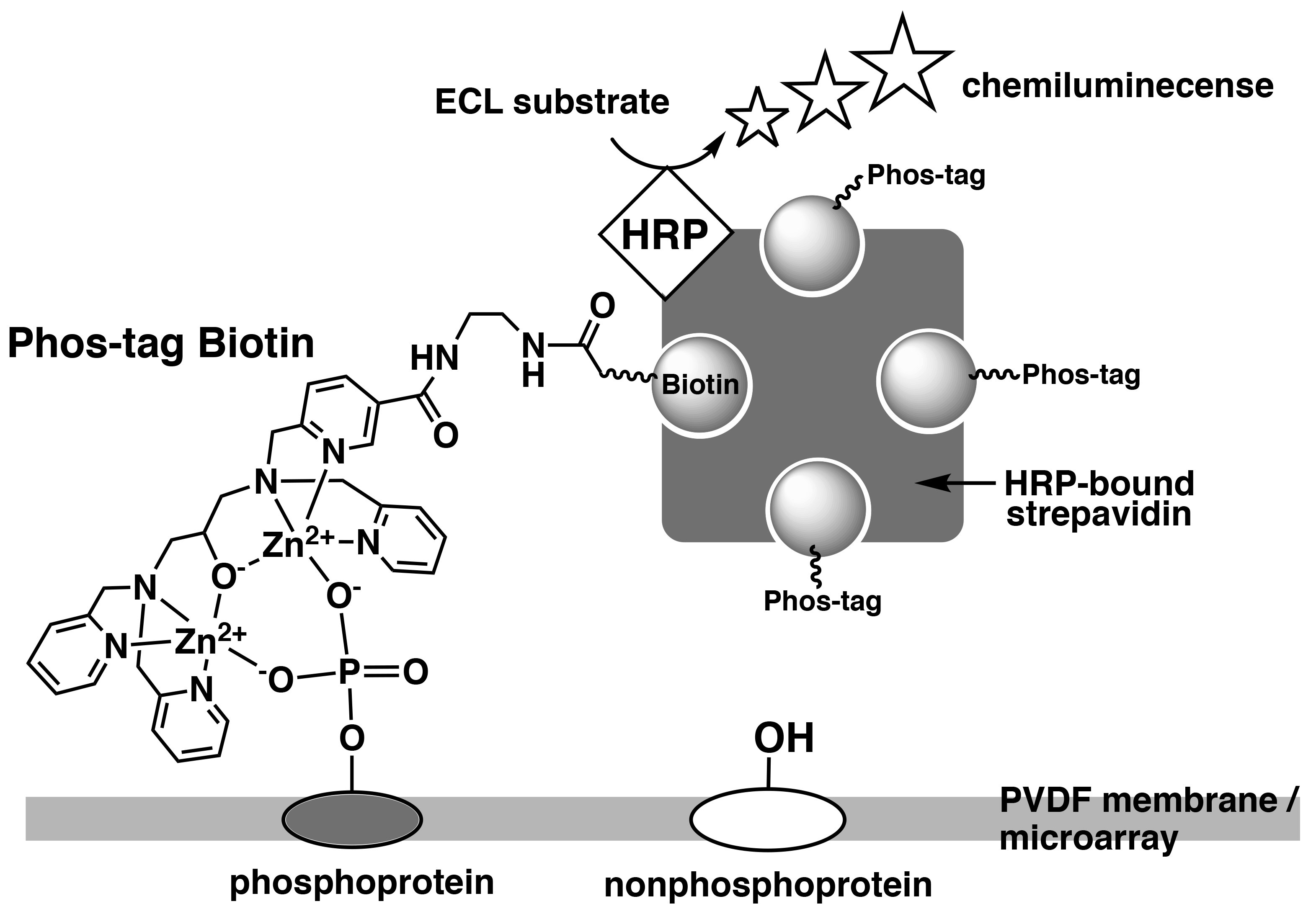
Figure 3.Detection of phosphorylated proteins on PVDF membrane or microarray using Phos-tag Biotin
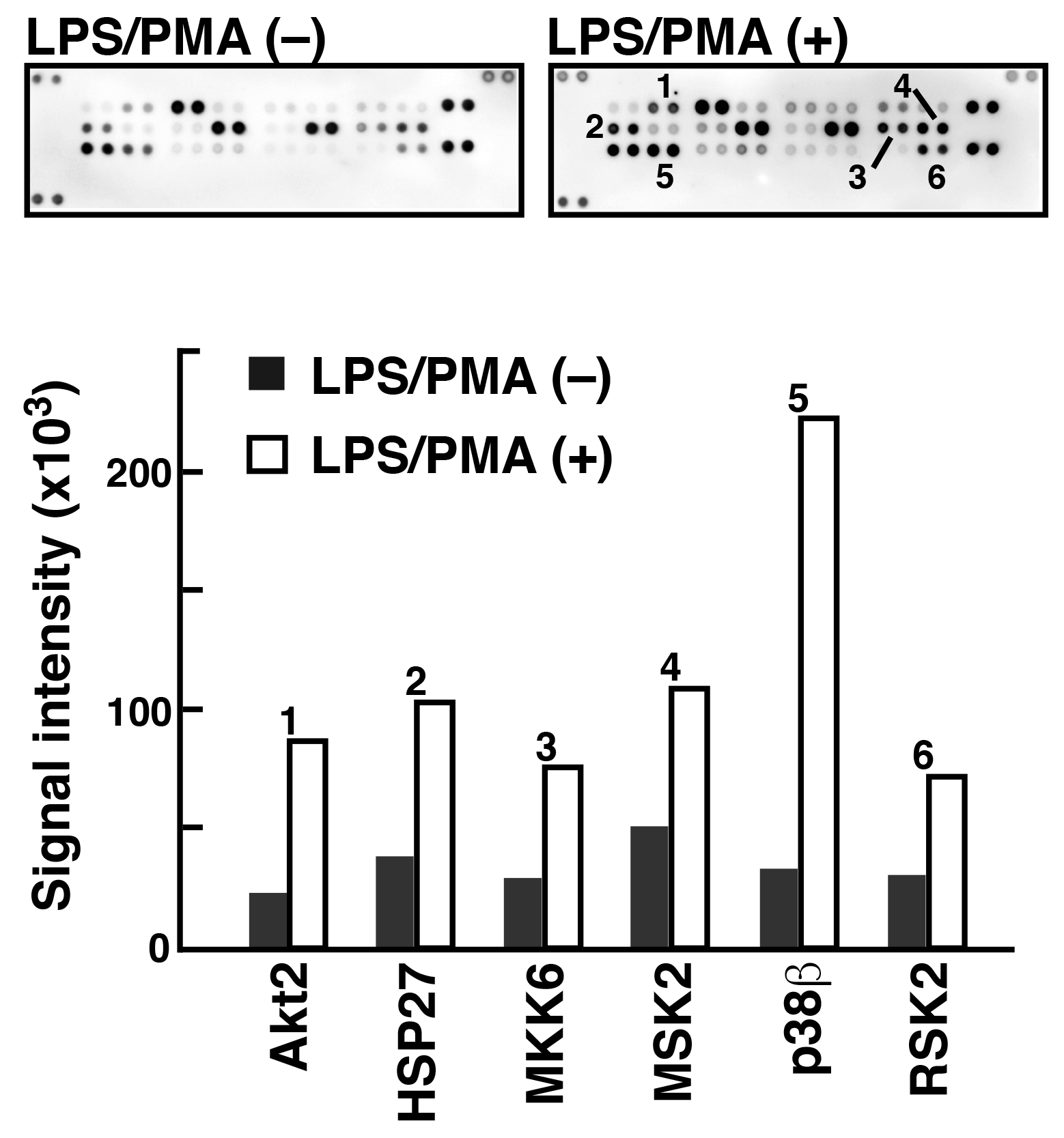
To analyze phosphorylated proteins using Phos-tag Biotin in conjunction with the ECL system, we used a commercially available antibody microarray Proteome Profiler (Human Phospho-MAPK Array, R&D Systems). 5) This microarray consists of 52 individual phosphorylation-independent capture antibodies spotted in duplicate on a nitrocellulose membrane. We examined the phosphorylated proteins present in two lysate samples of Raw 264.7 cells before and after treatment with lipopolysaccharide (LPS) and phorbol 12-myristate 13-acetate (PMA) (Figure 4). In agreement with the results of several other studies on LPS/PMA signaling, we confirmed the significant increase in the levels of phosphorylation of several proteins, including Akt2, HSP27, and p38b, using the antibody microarray system with Phos-tag Biotin. This technique allows simultaneous detection of the phosphorylation status of multiple proteins in a lysate sample.
Figure 4.Detection of phosphorylated proteins in Raw264.7 cells before and after the stimulation with LPS/PMA (reprint from reference 6)
An antibody microarray (Proteome Profiler Human Phospho-MAPK Array, R&D Systems) and Phos-tag Biotin were used to detect phosphorylated proteins in the Raw264.7 cell extract stimulated with LPS/PMA. Spots 1-6 showed significant phosphorylation after stimulation.
Separation and enrichment of phosphorylated proteins using Phos-tag Agarose
Phos-tag Agarose can be used as a matrix for phosphate-affinity chromatography. Although over 60% of all cellular proteins can be phosphorylated, they dynamically change between phosphorylation and dephosphorylation states using kinase and phosphatase reactions, respectively. In fact, the number of proteins in the phosphorylated state is very small. Therefore, the technique of separation and enrichment is very attractive and useful for the analysis of such phosphorylated proteins.7)
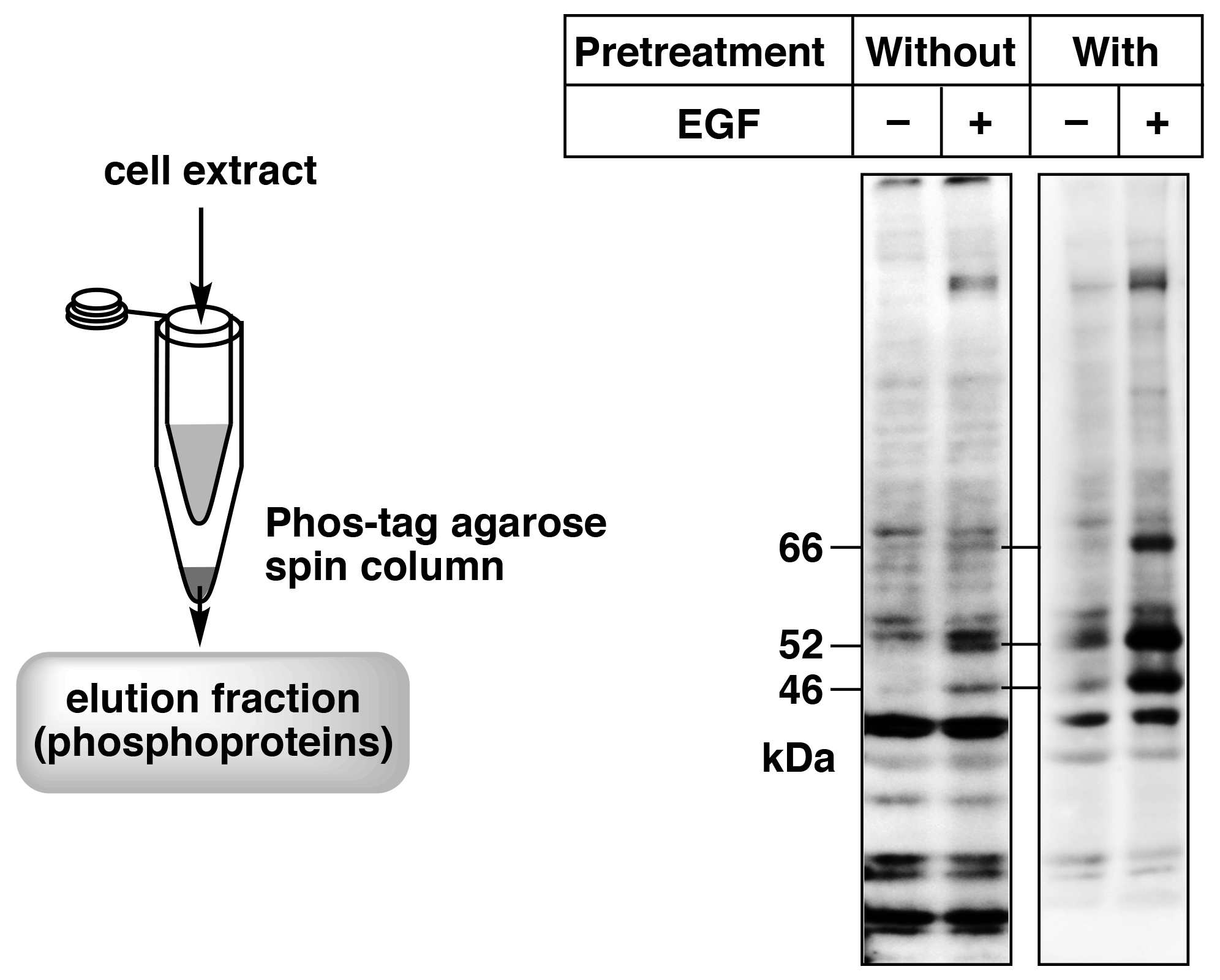
Figure 5. Enrichment of phosphorylated proteins by pretreatment of the cell extract samples using a simple spin column containing Phos-tag Agarose (reprint from reference 8) A431cells were stimulated with EGF, and SHC phosphorylation was analyzed by immunoblotting with a site-specific anti-phosphorylated SHC antibody. Pretreatment with a simple spin column containing Phos-tag Agarose enabled clear detection of the target bands (66, 52, and 46 kDa).
We have used Phos-tag Agarose in a spin column method to analyze phosphorylated proteins.8) In this method, a simple spin column is prepared using a 0.5-mL and a 1.5-mL microtube, and the column is packed with 20 μL Phos-tag Agarose (40 μL of Phos-tag Agarose slurry). Figure 5 shows the simple spin column and an immunoblot with a site-specific anti-phosphoprotein antibody against SHC of the cell extract without and with pretreatment with the simple spin column. The cell extract samples without pretreatment showed weak signals or nonspecific signals. Therefore, the phosphorylation states of SHC after stimulation by epidermal growth factor (EGF) could not be determined. On the other hand, samples pretreated with Phos-tag Agarose showed distinct signals, indicating an increase in phosphorylated SHC levels caused by EGF stimulation. These results demonstrate that pretreatment of cell extract with the simple spin column using Phos-tag Agarose can dramatically improve immunoblotting of phosphorylated proteins with site-specific anti-phosphoprotein antibodies. We therefore believe that use of a simple spin column with Phos-tag Agarose is a powerful tool for obtaining highly accurate data when using anti-phosphoprotein antibodies.
Phos-tag Agarose is also useful for the separation and enrichment of phosphopeptides from peptides mixtures in phosphoproteomics using mass spectrometry.9)
Conclusion
The number of research papers citing the use of Phos-tag was approximately 500 in 2018 (based on Google Scholar results), indicating that Phos-tag is gaining interest as a standard tool for the analysis of protein phosphorylation. It is noteworthy that Phos-tag-based techniques are used not only for the analysis of phosphoproteins in general life science research, but also for the analysis of bacterial or plant phosphoproteins that have been reported in relatively few studies. Furthermore, these techniques can be applied to analyze proteins for which antibodies are not commercially available as well as to proteins for which the phosphorylation status is unknown. We hope that more researchers use Phos-tag-based techniques that contribute to diverse areas of life science.
References
- Kinoshita, E. et al. : Mol. Cell. Proteomics, 5, 749 (2006).
- Kinoshita, E. and Kinoshita-Kikuta, E. : Proteomics, 11, 319 (2011).
- Kinoshita-Kikuta, E. et al. : PLoS One, 10, e0132598 (2015).
- Kinoshita-Kikuta, E. et al. : Int. J. Chem., 4, e1 (2012).
- Kinoshita-Kikuta, E. et al. : Biochim. Biophys. Acta Proteins Proteom., 1867, 62 (2019).
- Kinoshita, E. et al. : Anal. Biochem., 438, 104 (2013).
- Kinoshita-Kikuta, E. et al. : Proteomics, 6, 5088 (2006).
- Kinoshita-Kikuta, E. et al. : Anal. Biochem., 389, 83 (2009).
- Yuan, E. T. et al. : Electrophoresis, 38, 2447 (2017).




