Restriction Enzymes / Modifying Enzymes
Restriction enzymes recognize and cleave specific sequences of double-stranded DNA. The use of restriction enzymes made DNA analysis and recombinant DNA technology, and contributed greatly to the advances in life science research. Fujifilm Wako offers a wide range of restriction and modification enzymes.
Product Line-up
More Information
Cloning and structural/functional analysis of genes involve procedures such as digestion, modification, and degradation of nucleic acids. Various enzymes are used for these purposes: restriction enzymes for nucleic acid cleavage, phosphatases/kinases for modifications, nucleases for degradation, and so forth. An overview of these enzymes is presented and properties are described below.
Restriction Enzymes
Restriction enzymes recognize and cleave specific sequences (recognition sequences) in double-stranded DNA. These enzymes are used to prepare DNA fragments to be inserted into a vector in the process of cloning. Restriction enzymes are originally used by bacteria to cleave foreign DNA to protect themselves from infection by phages. (The bacteria modify their own DNA to prevent it from being cleaved by the restriction enzyme.)
Restriction enzymes are classified into four types according to the mode of cleavage and the factors required for the reaction.
Type I
Type I restriction enzymes consist of three different subunits (sequence recognition, endonuclease, and methyltransferase subunits). The cleavage site is separated from the recognition sequence by various lengths that are at least 1 kb. The reaction requires S-adenosylmethionine (SAM) and ATP.
Type II
Type II restriction enzymes have sequence recognition capability and endonuclease activity. They do not have modifying activity. They are the main enzymes used for cloning, because they precisely cleave the recognition site (or its vicinity). Mg2+ is required for the reaction.
Type III
Type III restriction enzymes consist of two subunits. The cleavage site is separated from the recognition sequence by about 25 to 28 bp. The reaction requires ATP and Mg2+.
Type IV
Type IV restriction enzymes cleave methylated, hydroxymethylated, or glucosylhydroxymethylated DNA. The recognition and cleavage sites are separated.
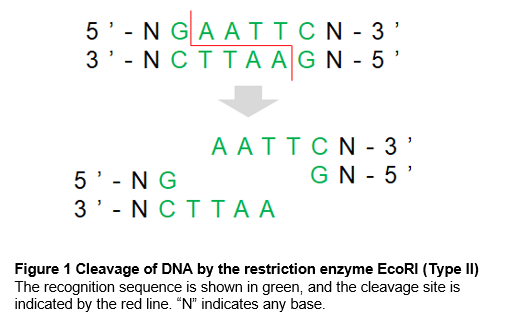
Restriction enzymes have unique recognition sequences, many of which are palindromic (Figure 1). When different restriction enzymes recognize the same sequence, they are called isoschizomers. When they recognize the same sequence but cleave in different positions, they are called neoschizomers.
Restriction enzymes differ not only in the recognition sequence, but also in the cleavage pattern. The cleavage patterns are broadly classified into “sticky ends” (5' or 3' protruding ends) and “blunt ends.” In general, sticky ends have a better efficiency for subsequent ligation than the blunt ends.
Restriction digestion is usually performed at 37°C. Since the optimal salt concentration for the reaction varies with the restriction enzyme, an appropriate buffer for each restriction enzyme must be selected.
Modification Enzymes
Enzymatic modifications of nucleic acids include dephosphorylation and phosphorylation.
The ligation reaction of double-stranded DNA joins the phosphate group at the 5' end with the hydroxyl group at the 3' end. Therefore, phosphorylation and dephosphorylation are important pretreatments for the ligation reaction. Dephosphorylation of the 5' end is effective in preventing self-ligation of plasmid vectors. Bacterial alkaline phosphatase (BAP) and calf intestinal alkaline phosphatase (CIP) are commonly used for dephosphorylation. On the other hand, phosphorylation of DNA fragments is required when fragments without 5' phosphate are used for ligation. Phage-derived T4 polynucleotide kinase is used for phosphorylation.
Nucleases
Enzymes that degrade both DNA and RNA are called “nucleases.” Exonucleases degrade nucleic acid chains from the ends, and endonucleases cleave nucleic acids in the middle of chains. Restriction enzymes are a type of endonuclease. DNases cleave only DNA, and RNases cleave only RNA. These enzymes are used to remove DNA and RNA in the process of nucleic acid extraction, to make blunt-ended DNA fragments, and to study structures and interactions of DNA/RNA. Below are properties and applications of typical nucleases.
| Nuclease | Substrate | Single-stranded | Double-stranded | Properties | Applications |
|---|---|---|---|---|---|
| DNase I | DNA | ○ | ○ | Non-specifically degrades DNA in the presence of Mg2+. | Removal of DNA, DNase footprint analysis, etc. |
| RNase A | RNA | ○ | × | Cleaves the phosphodiester bonds at the 3' side of the pyrimidine base. | Removal of RNA, etc. |
| RNase H | RNA | × | ○ | Degrades RNA in DNA/RNA hybrid chains. | Degradation of template RNA after reverse transcription, etc. |
| Nuclease P1 | DNA RNA |
○ | × | Works on single-stranded portions of double-stranded nucleic acids. The reaction proceeds in the presence of Zn2+. | Selective degradation of single-stranded DNA, generation of blunt-ended DNA fragments, etc. |
| Nuclease S1 | DNA RNA |
○ | × | Works on single-stranded portions of double-stranded nucleic acids. The reaction proceeds in the presence of Zn2+. | Selective degradation of single-stranded DNA, generation of blunt-ended DNA fragments, S1 mapping, etc. |
| Mung Bean Nuclease | DNA RNA |
○ | × | Works on single-stranded portions of double-stranded nucleic acids. It has a higher propensity to generate blunt ends compared to S1 nuclease. | Selective degradation of single-stranded DNA, generation of blunt-ended DNA fragments, etc. |
References
- Roberts, R. J. et al.: Nucleic Acids Res., 31(7), 1805(2003)
A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes - “Biochemical dictionary 4th ed.” ed. by Oshima, T. et al., Tokyo Kagaku Dojin, Japan, (2007). (Japanese)
- “Genetic engineering experiment notebook Vol. 1 3rd ed.” ed. by Tamura, T., Yodosha, Japan, (2010). (Japanese)
- Green, R. M. and Sambrook, J.: ”Molecular Cloning A Laboratory Manual, 4th ed.”, Cold Spring Harbor Laboratory Press, (2012).
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.



