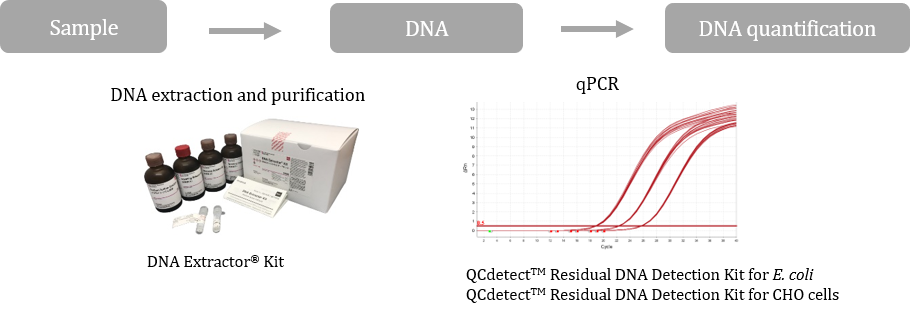
Test kits for residual DNA from host cells
A CHO cell or Escherichia coli is used as a host cell for expression of antibodies or proteins in the manufacturing process of biopharmaceuticals. As DNA of these host cells may cause tumor formation, it is necessary to control the DNA as a process-related impurity.
Guidelines of the World Health Organization (WHO), the United States Food and Drug Administration (USFDA), and the European Pharmacopoeia (EP) state that the final amount of host residual DNA should be less than 100 pg/dose, or even as little as 10 ng/dose.
FUJIFILM Wako Pure Chemical Corporation offers a DNA extraction and purification kit, "DNA Extractor® Kit," as a reagent for residual DNA testing. In addition, we have newly released residual DNA Detection Kit for E. coli and CHO cells.
QCdetect™ Residual DNA Detection Kit for E. coli
This product is a kit for quantifying genomic DNA from E. coli by qPCR (probe method).
It can be used for residual DNA testing for biopharmaceuticals and their manufacturing processes.
- Limit of detection : ≥ 0.03 pg/test
- Limit of quantitation : ≥ 0.3 pg/test
- Calibration curve with high linearity
- Minimal inter-assay variability and high reproducibility
- Easy operation using the pre-mix buffer
- Less susceptible to impurities in sample
- Contains an internal control
QCdetect™ Residual DNA Detection Kit for CHO cells
This product is a kit for quantifying genomic DNA from CHO cells by qPCR (probe method).
It can be used for residual DNA testing for antibody pharmaceuticals and their manufacturing processes.

- Limit of detection: ≥ 0.0003 pg/test
- Limit of quantitation: ≥ 0.003 pg/test
- Calibration curve with high linearity
- Minimal inter-assay variability and high reproducibility
- Easy operation using the pre-mix buffer
- Less susceptible to contaminants in samples
- Contains an internal control
DNA Extractor® Kit
A kit for extracting residual DNA from host cells by sodium iodide method. The extracted DNA can be quantified by qPCR.
This kit can be used for testing and control of the quantity of residual DNA obtained from host cells such as CHO cells, Escherichia coli, and yeast.
- Trace amounts of DNA (100-1,000 fg) can be recovered in high yield
- No need for tube replacement
(all the processes can be completed in the same tube) - Only 60-90 minutes needed from start to completion of extraction
- A protocol for high-protein samples is available
- Sodium iodide method* is adopted
*The sodium iodide method is a residual DNA extraction technique that is described in the United States Pharmacopeia (USP) 42-NF37, <509> “Residual DNA Testing”.
Product List
- Open All
- Close All
QCdetect™ Residual DNA Detection Kit for E. coli
QCdetect™ Residual DNA Detection Kit for CHO cells
DNA Extractor® Kit
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.



