Isothermal Amplification Reagents
A gene amplification method using thermostable strand-displacement DNA polymerase, called the loop-mediated isothermal amplification (LAMP) method, is a gene amplification technology that is completely different from PCR. The LAMP method requires no dedicated device such as a thermal cycler and can amplify DNA with a heat block or water bath. Fujifilm Wako offers LAMP reagents, including thermostable strand-displacement DNA polymerases.
More Information
DNA Amplification Methods

PCR is the most common method used for DNA amplification. It is used to test for the presence of target DNA or to determine the quantity of target DNA because of its ability to amplify DNA 2n times. However, special equipment (thermal cycler) is required to precisely raise and lower the temperature, and when the sequence has a high GC content, DNA synthesis may be inhibited by a secondary structure of DNA. Accordingly, a gene amplification method using thermostable strand-displacement DNA polymerase, called the loop-mediated isothermal amplification (LAMP) method*, was developed.
* LAMP is a patented technology belonging to Eiken Chemical Co., Ltd
Comparison of LAMP and PCR methods
| LAMP method | PCR method | |
|---|---|---|
| Temperature | Constant (60 – 65°C) | Requires thermal denaturation at high temperature (95°C) |
| Time | 15–60 min. | 2–3 hours |
| Amount of product | 1010✖ | 107✖ |
| Specificity | Extremely high 6 regions 4 primers |
High 2 regions 2 primers |
| Detection | Presence or absence of amplification by-product | Requires a separate detection step |
LAMP Method
In LAMP analysis, the presence or absence of a target DNA sequence can be determined based on turbidity, fluorescence, color development, or color change. As DNA amplification can be performed at isothermal temperatures and can be confirmed visually without analysis such as electrophoresis, it is expected that the LAMP method will be applied to genetic testing outside of laboratories or in developing countries.
▼Principles
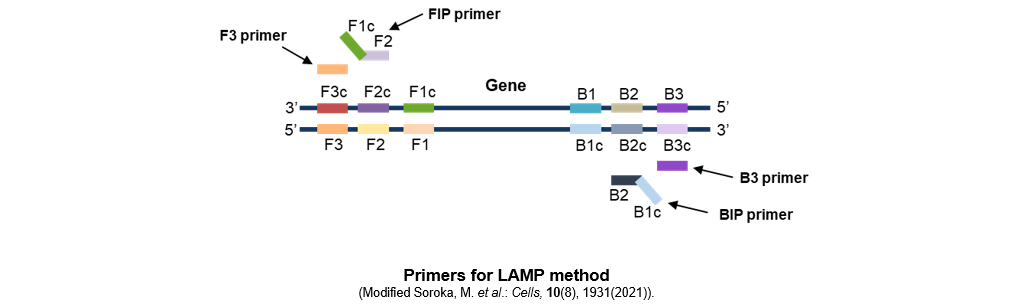
In the LAMP method, six distinct regions are set on both sides of the target DNA (three on the F side and three on the B side) and four different primers (F3, FIP, B3, BIP) are used. Using thermostable strand-displacement DNA polymerase, loop structures are formed at both ends of the target sequence and the target sequences are linked together, which enables a large number of repeating structures to be formed in a short time. Additionally, as the thermostable strand-displacement DNA polymerase synthesizes a new DNA strand while dissociating the hydrogen bond of the double-stranded template DNA, the amplification of nucleic acids proceeds at a constant temperature.
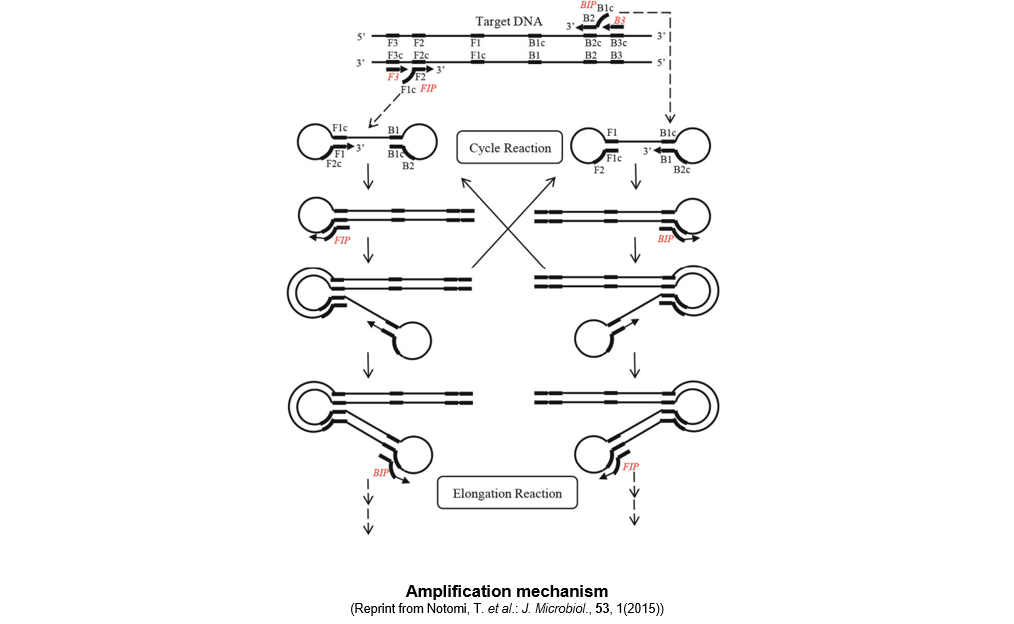
▼Amplification mechanism
When a reaction reagent containing the thermostable strand-displacement DNA polymerase, primers, and a DNA sample are allowed to react at a constant temperature of 60 to 65°C, the amplification reaction proceeds as described below.
At a temperature between 60 and 65°C, double-stranded DNA is in a dynamic state, resulting in the appearance of single-stranded DNA regions. A primer anneals to the region and initiates DNA synthesis, which in turn leads to dissociation of double-stranded DNA into single-stranded DNA. A starting structure for the LAMP amplification cycle is generated from the single-stranded DNA. [step 1) to 4)]
- The FIP primer anneals to the single-stranded DNA and a DNA strand complementary to the template DNA is synthesized.
- The F3 primer, designed to anneal to a region outside of that for the FIP primer, anneals to the DNA synthesized in step 1) and initiates synthesis while the double-stranded DNA is being dissociated. During synthesis, a single-stranded DNA is released, which has two complementary regions (F1c and F1) at the 5’ end and forms a loop structure by self-annealing.
- The BIP primer anneals to the 3’ end of the single-stranded DNA with the loop structure at the 5’end and initiates DNA synthesis.
- The B3 primer, designed to anneal to a region outside of that for the BIP primer, anneals to the DNA synthesized in step 3) and initiates synthesis while the double-stranded DNA is being dissociated. During synthesis, a single-stranded DNA is released, which has F1c and F1 at the 5’ end and B1c and B1 at the 3’ end and both ends form a loop structure by self-annealing. This structure serves as the starting structure for the amplification cycle of the LAMP method.

After formation of the starting structure, the reaction proceeds as described below.
▼Features
High specificity
As the LAMP method requires six gene regions and four primers, it has superior specificity compared to PCR. The presence of the target gene can be determined by detecting the amplified product.
High amplification efficiency
The LAMP method does not require thermal denaturation (dissociation of double-stranded DNA), therefore the synthesis is less susceptible to the inhibitory effect of secondary DNA structures. In addition, its amplification principle enables detection within one hour.
Easy detection
Because of the large amounts of amplification products, when the amplification process is performed in the presence of a fluorescent dye that binds to a double-stranded DNA, the amplified product can be confirmed visually under an ultraviolet light lamp. It is known that pyrophosphoric acid generated as a by-product of DNA synthesis binds to a magnesium ion in the reaction solution. When magnesium pyrophosphate is produced in large quantities by the amplification reaction and the value of the solubility product is exceeded, a white precipitate appears, which can be used as an indicator for amplification and can be detected visually or with turbidity measuring equipment. Detection by real-time PCR is also possible.
▼Basic procedures
1. Preparation of equipment and reagents
Equipment
At present, two main types of detection equipment are commercially available.
(A) Turbidity measuring equipment for measurement of the turbidity of magnesium pyrophosphate
(B) Fluorescence detector for measurement of fluorescent molecules that bind to double-stranded DNA
Reagents
Basic reagents are as follows:
- Thermostable strand-displacement DNA polymerase
- dNTPs
- Reaction buffer
- Four types of primers
2. Preparation of primers
In the LAMP method, primer sets designed to be sequence-specific or customized primer sets are used. Primers are designed as follows:
- Define 6 regions (B1, B2, and B3 that are upstream of the target gene and F1c, F2c, and F3c that are downstream of the target gene).
- Design 4 primers (FIP, F3, BIP, and B3) for the 6 regions.
FIP (Forward Inner Primer)
FIP is designed to include the same sequence as the F1c region at the 5' end and the F2 sequence that is complementary to the F2c region at the 3' end.
F3
F3 is designed to include a sequence that is complementary to the F3c region.
BIP (Backward Inner Primer)
BIP is designed to include the same sequence as the B1c region at the 5' end and the B2 sequence that is complementary to the B2c region at the 3' end.
B3
B3 is designed to include a sequence that is complementary to the B3c region.
3. Conduct and detection of LAMP reaction
DNA amplification can be detected by turbidity, fluorescence, or visual inspection.
Turbidity detection: The turbidity of magnesium pyrophosphate produced by the amplification reaction is measured.
Fluorescence detection: A fluorescent dye is intercalated into DNA and detected by a fluorescence detection device such as a real-time PCR detection system.
Visual detection: DNA amplification is detected visually by adding fluorescent reagents for visual detection.
DNA Amplification Methods
Fujifilm Wako supplies a line-up of three types of thermostable strand-displacement DNA polymerase from NIPPON GENE that can be used for LAMP. Among them, Bst DNA polymerase is the most frequently used enzyme. In addition, Fujifilm Wako provides Csa DNA polymerase and 96-7 DNA polymerase, with different reaction and deactivation temperatures to enable selection of an enzyme suitable for your experimental system.
Comparison of Three strand-displacement DNA polymerases
| Enzyme name | Bst DNA Polymerase | Csa DNA Polymerase | 96-7 DNA Polymerase |
|---|---|---|---|
| Reaction temperature | 60-65℃ | 60-70℃ | 50-55℃ |
| Deactivation temperature | 80°C for 5 min. | 85°C for 5 min. | 70°C for 5 min. |
References
- Notomi, T. et al.: Nucleic Acids Res., 28(12), E63(2000).
Loop-mediated isothermal amplification of DNA - Soroka, M., Wasowicz, B. and Rymaszewska, A.: Cells, 10(8), 1931(2021).
Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? - Notomi, T., Mori, Y., Tomita, N. and Kanda, H.: J. Microbiol., 53(1), 1(2015).
Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects - Ushikubo, H.: Uirusu, 54(1),107(2004).
Principle of LAMP method - a simple and rapid gene amplification method - (Japanese) - Eiken GENOME SITE: https://loopamp.eiken.co.jp/lamp/ (Japanese) (Nov. 7th, 2022)
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.
The prices are list prices in Japan.Please contact your local distributor for your retail price in your region.



