RiboNAT™ Rapid Sterility Test - RNA Isolation Kit 1
- for Microorganism Detection
- Manufacturer :
- FUJIFILM Wako Pure Chemical Corporation
- Storage Condition :
- Keep at -20 degrees C.
- GHS :
-
- Structural Formula
- Label
- Packing
- SDS
|
Comparison
|
Product Number
|
Package Size
|
Price
|
Inventory
|
|
|---|---|---|---|---|---|
|
|
|
50Tests
|
|
In stock in Japan |
Please check here for notes on products and prices.
Document
Kit component
50 Tests
| Proteinase K Solution | 700 μL ×1 |
|---|---|
| Enzyme Mix | 750 μL ×1 |
| DNase Solution | 380 μL ×1 |
| 1st-DNase Buffer (1) | 23 mL ×1 |
| 2nd-DNase Buffer (2) | 180 μL ×1 |
| Activator Solution 1 (SCDM) | 50 mL ×1 |
| Activator Solution 2 (TG) | 50 mL ×2 |
| Enzyme Enhancer | 50 mL ×1 |
| Nucleic Acid Inactivator | 60 μL ×1 |
Overview
As part of safety testing for pharmaceuticals, sterility tests are conducted to check for microbial contamination. The compendial sterility test method requires a 14-day incubation period, which has increased the demand for faster testing methods, especially for cell-based medicines with short shelf lives. RiboNAT™ utilizes the NAT method (Nucleic Acid Amplification Test) to rapidly detect bacteria and fungi.
Features
- Short time (7 hours) and high sensitivity (9 CFU/mL).
- Extracts total RNA of microorganisms and detects ribosomal RNA.
- Wide range of bacteria and fungi can be detected with a single assay (qualitative test).
- Reduction of false positives from dead microorganisms and residual DNA.
Assay Flow
- Step 1. Pre-treatment
-
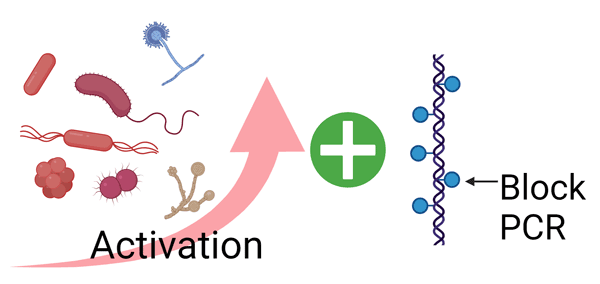
- Activation of microorganisms.
- Inactivation of residual DNA.
3.5 hours- RNA Isolation Kit 1
- Step 2. RNA isolation
-
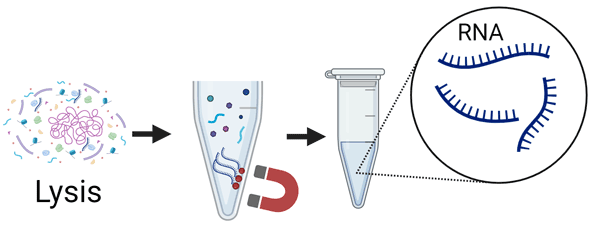
- Lysis of microorganism.
- RNA extraction & purification.
1.5 hours- RNA Isolation Kit 1
- RNA Isolation Kit 2
- Detection Kit
- Step 3. Measurement
-
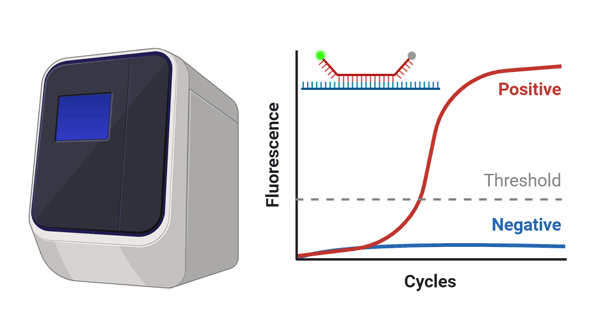
- RNA detection with reverse
transcription real time PCR.
1.2 hours- RNA Isolation Kit 1
- Detection Kit
- RNA detection with reverse
*Illustrations were created with BioRender.
h
o
u
r
s
*RiboNAT™ RNA Isolation Kit 1, RNA Isolation Kit 2, Detection Kit are all required for the assay.
Specifications of Detection Kit
| Detection method | One step reverse transcription real time PCR with fluorescence probe (RT-rt PCR) |
|---|---|
| Sensitivity | 100 RNA copies per reaction |
| Coverage* | Bacteria: 25,748 (95.7%) Fungi: 1,683 (92.3%) *In silico analysis, Accepting 3 mismatches, Data bank: Silva |
| Target | Bacteria: 23S Ribosomal RNA (Detection wavelength: 515-530nm) Fungi: 25/28S Ribosomal RNA (Detection wavelength: 675-690 nm) Internal Control: Artifact sequence (Detection wavelength: 560-580 nm) |
Performance
Detection of 6 species described in pharmacopeias : Spiked 9 CFU/mL
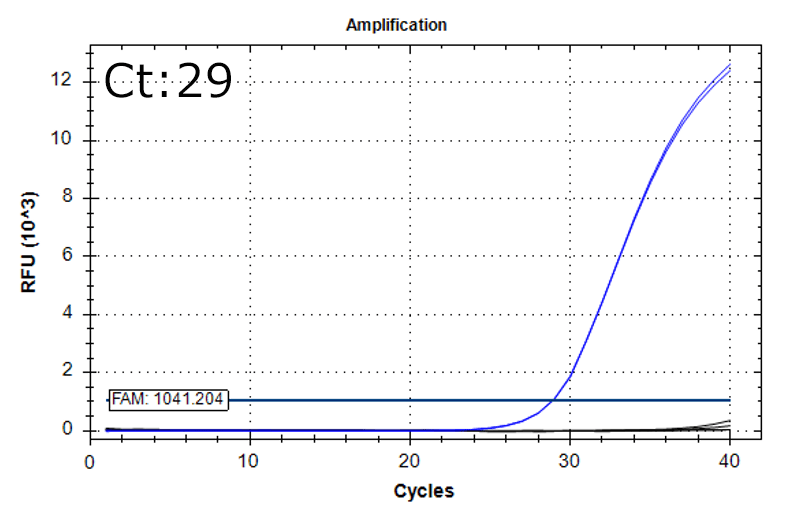
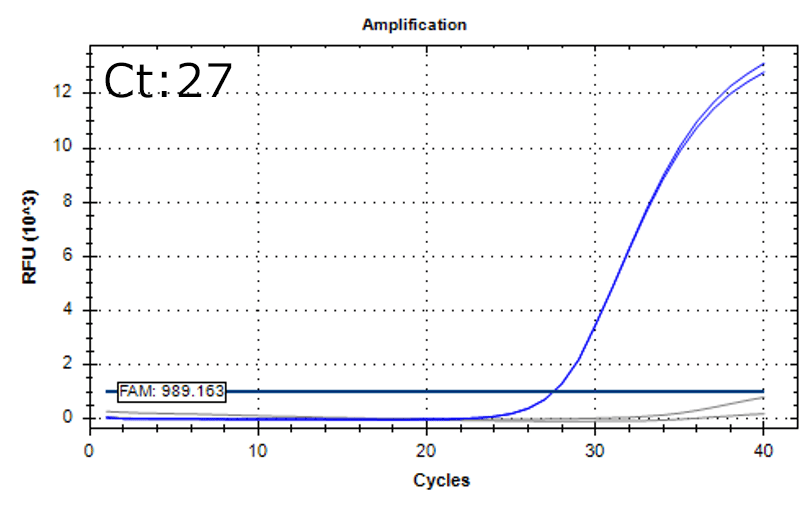
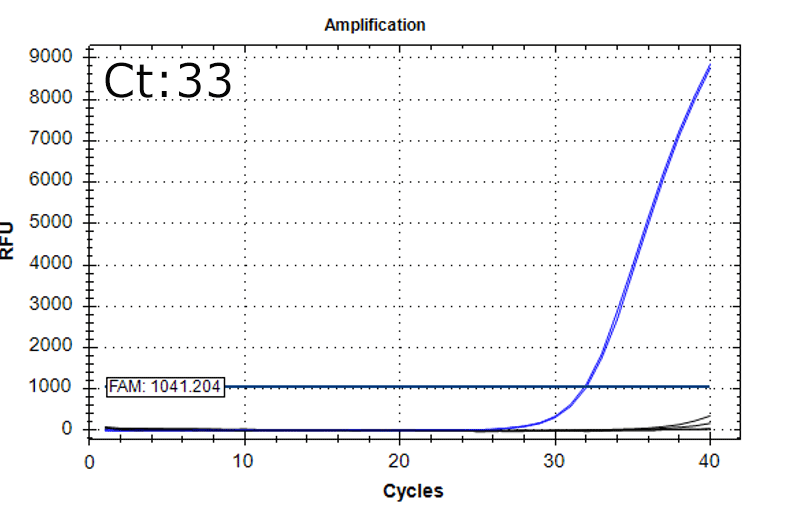
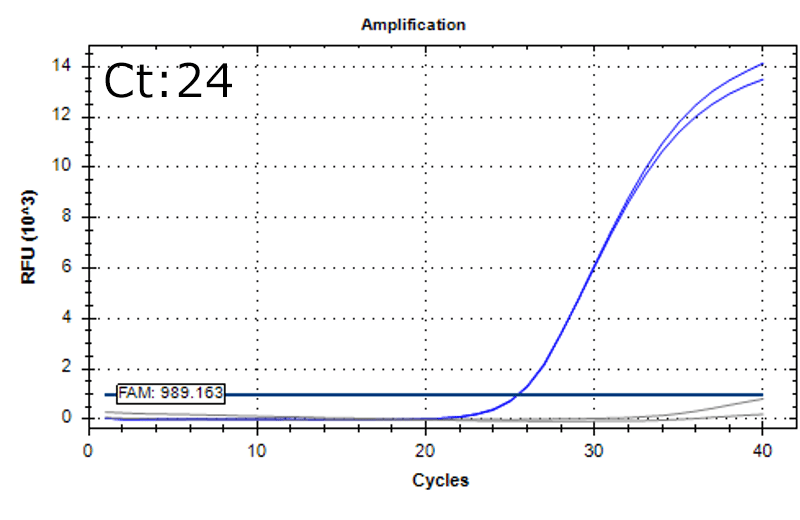
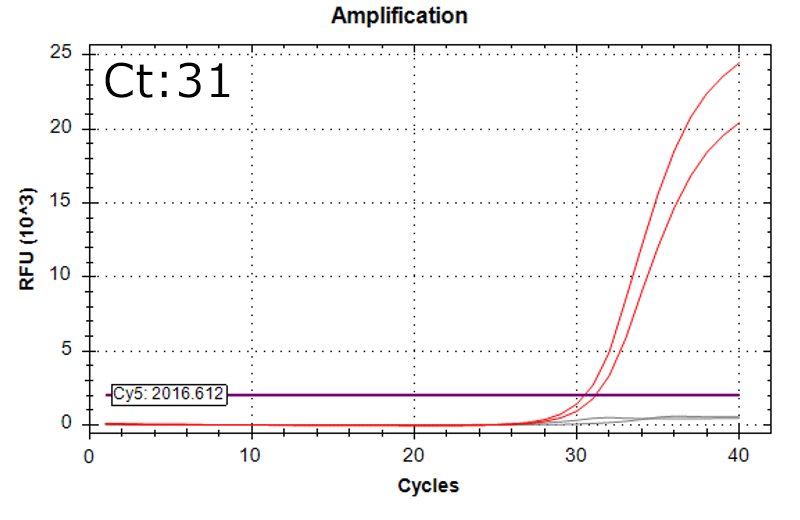
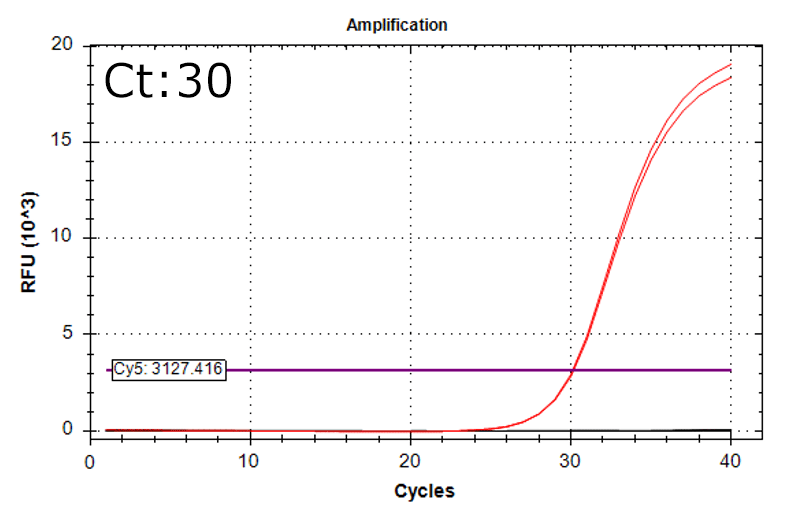
Detectable at 9 CFU/mL
Spiked 9 CFU/mL with cell suspension sample
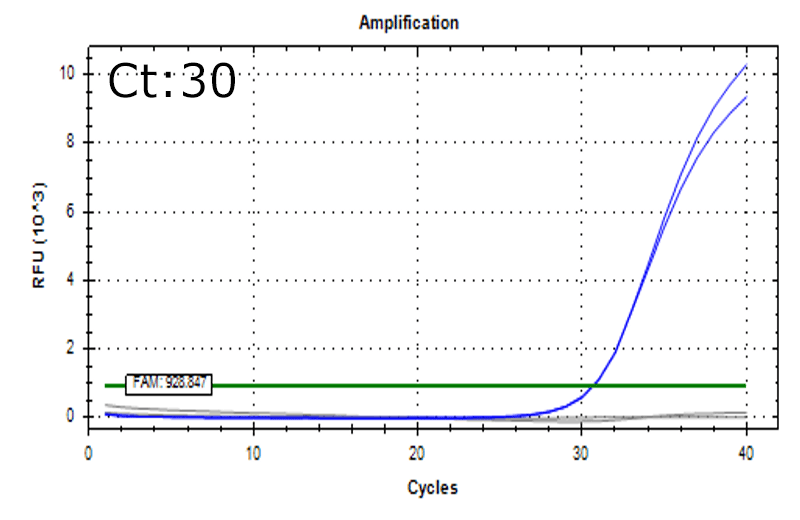
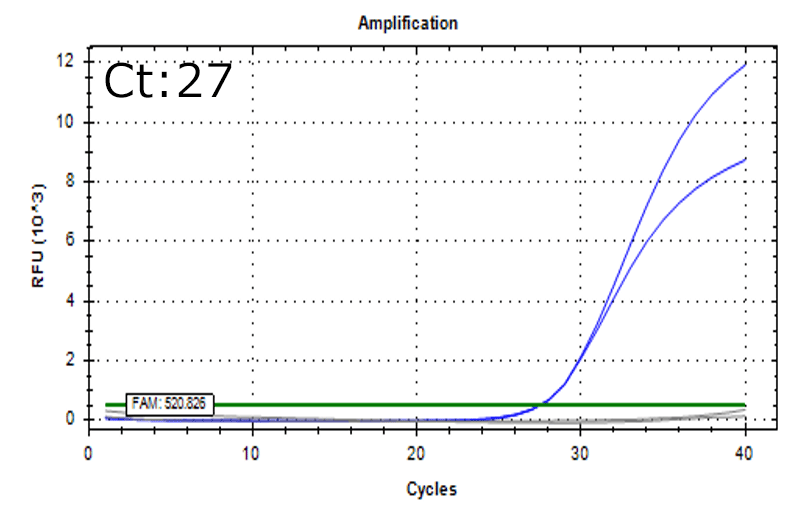
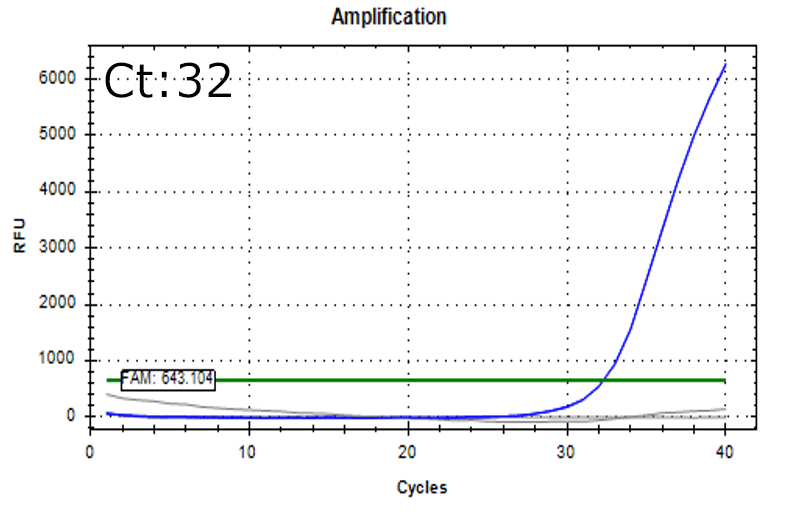
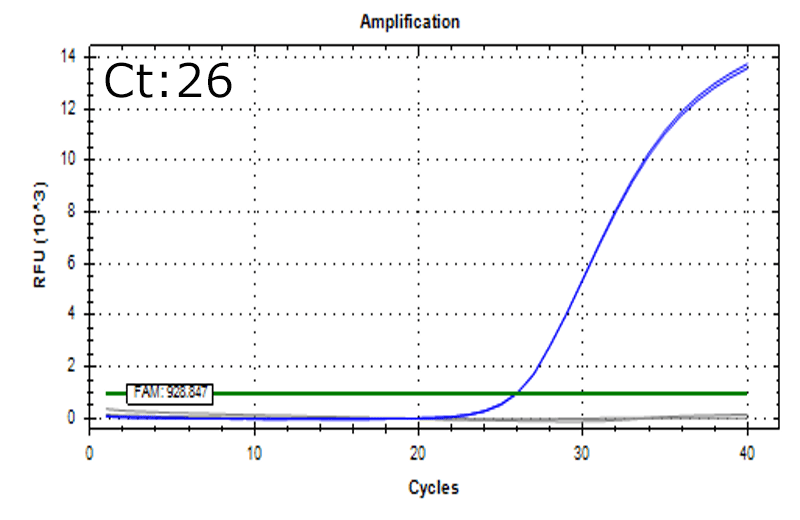
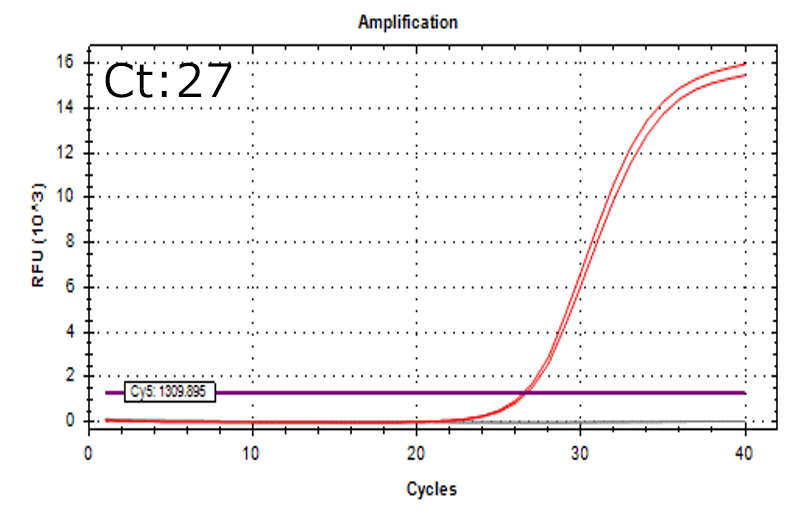
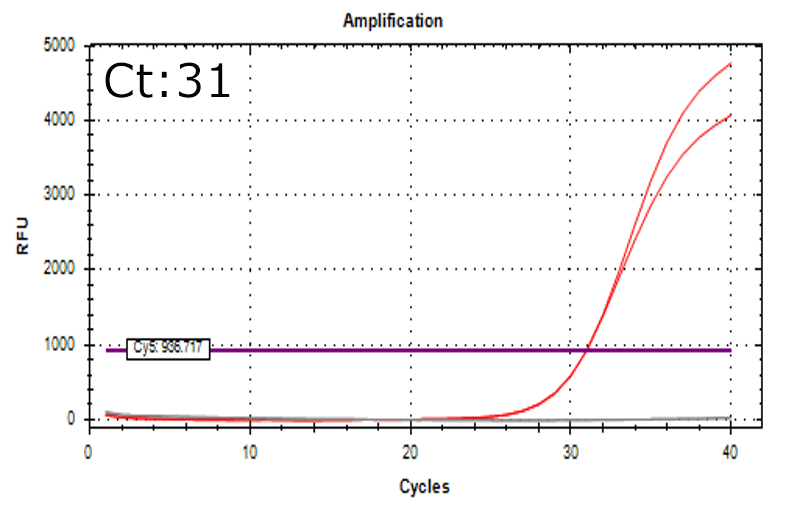
Test condition
- Cells: HEK293
- Concentration: 0.25 x 106 cells/mL
*Detectable also in the below condition
- Mesenchymal stem cells (MSC): 0.5 x 106 cells/mL
- T-cells:1.0 x 106 cells/mL
Detectable at 9 CFU/mL with cell suspension sample
Higher sensitivity method
- Inoculated strain: 2 CFU/mL
- Incubation time: 3-15 hours
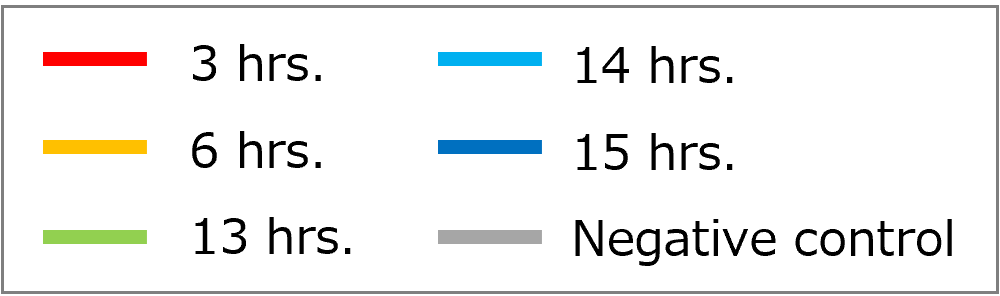
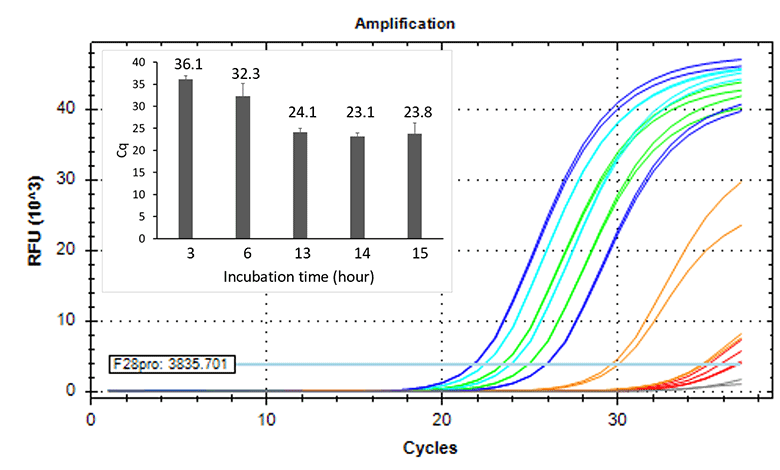
BIOBALL®
Strain: NCPF2275
< Fungi >
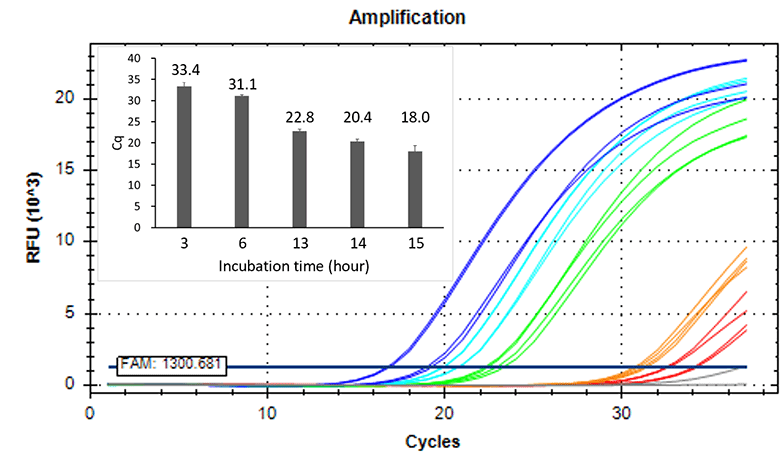
BIOBALL®
Strain: NCTC12935
< Strictly anaerobic bacteria >
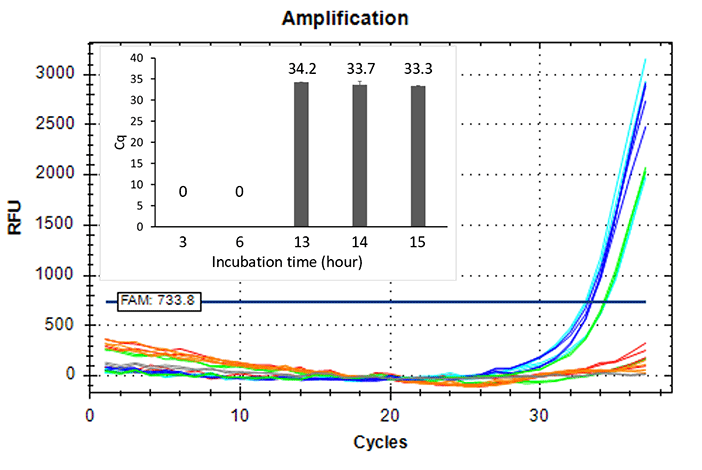
BIOBALL®
Strain: DSM1897
< Facultative anaerobic bacteria
Doubling time: 5 hrs. >
Detected 2 CFU/mL of strictly anaerobic bacteria and slow growth bacteria by 14 hours or longer incubation.
Extended incubation enhances sensitivity
Reduction of False Positive
RiboNAT™ includes treatment steps with a reagent for inactivating DNA from dead cells and DNase. This process helps reduce false positives caused by residual DNA in the sample.
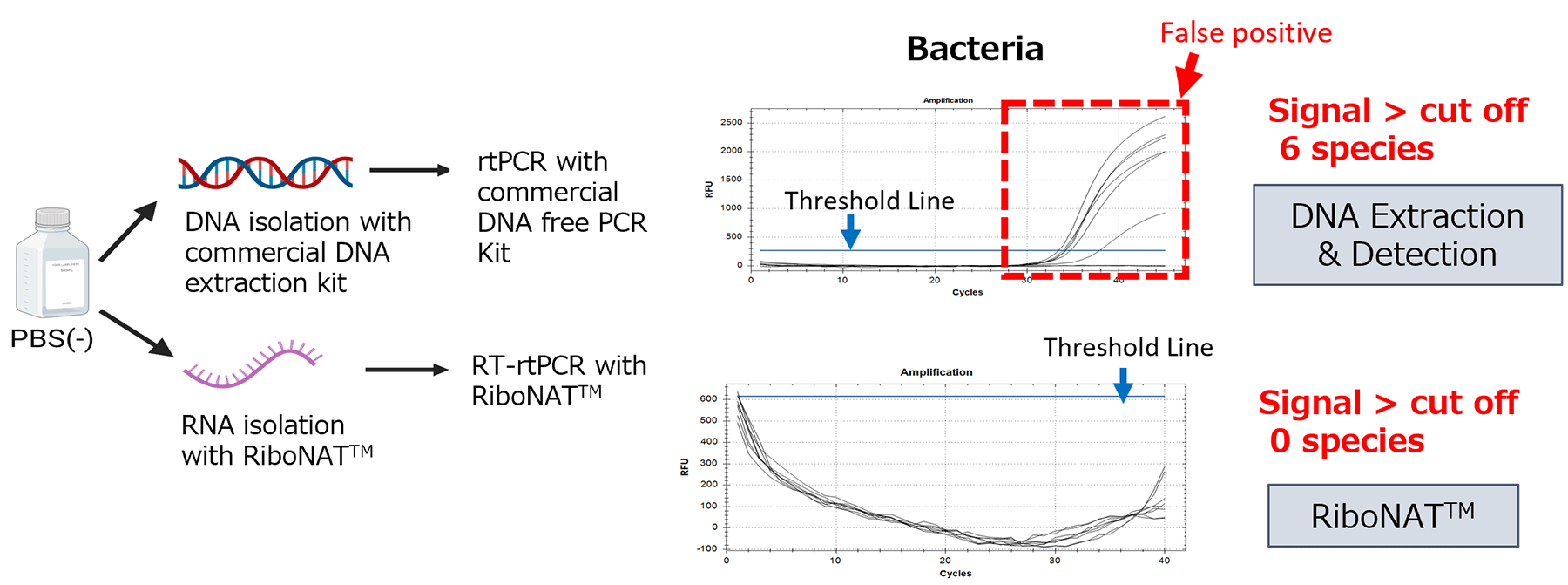
False positive derived from residual DNA was reduced
Instruction Video
FAQ
- What is the required sample volume for the test?
- According to the standard protocol, total 2 mL of the sample is centrifuged and used for the test. While testing with smaller sample volumes is possible, the likelihood of detecting microorganisms decreases because fewer microbes are present in the smaller volume of samples.
- How many samples can be measured with one kit?
- For each sample, RNA extraction is performed once, and PCR is conducted in duplicate wells (n=2) per extraction. In addition to samples, each measurement requires controls as follows: Negative Extraction Control (1 extraction → 2 PCR wells), No-Template Control (4 PCR wells), and PCR Positive Control (4 PCR wells).
The RNA Isolation Kits support 50 extractions, and the Detection Kit supports 100 PCR wells.
- Do reagents need to be used all at once?
- No. After use, please store them at the appropriate temperature and use them before the expiration date.
- Which real-time PCR instruments are compatible?
- The CFX96 System (Bio-Rad Laboratories, Inc.) and QuantStudio® 5 Real-Time PCR System (Thermo Fisher Scientific Inc.) have been validated. Other instruments capable of detecting the specified wavelengths may also be used.
Overview / Applications
Property
Manufacturer Information
Alias
For research use or further manufacturing use only. Not for use in diagnostic procedures.
Product content may differ from the actual image due to minor specification changes etc.
If the revision of product standards and packaging standards has been made, there is a case where the actual product specifications and images are different.
The prices are list prices in Japan.Please contact your local distributor for your retail price in your region.





